6IEN
 
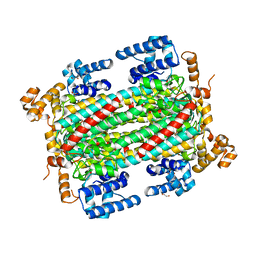 | | Substrate/product bound Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ARGININOSUCCINATE, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
6IEM
 
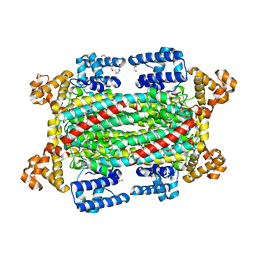 | | Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, Argininosuccinate lyase, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
2HV8
 
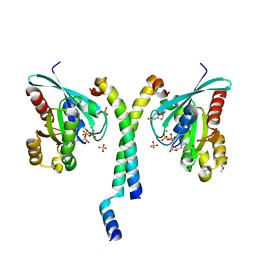 | | Crystal structure of GTP-bound Rab11 in complex with FIP3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eathiraj, S, Mishra, A, Prekeris, R, Lambright, D.G. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Rab11-mediated Recruitment of FIP3 to Recycling Endosomes.
J.Mol.Biol., 364, 2006
|
|
4LQ6
 
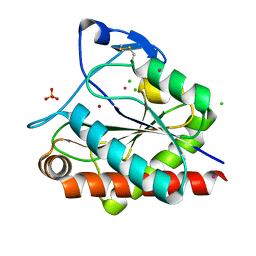 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3MJH
 
 | | Crystal Structure of Human Rab5A in complex with the C2H2 Zinc Finger of EEA1 | | Descriptor: | Early endosome antigen 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mishra, A.K, Eathiraj, S, Lambright, D.G. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6YAU
 
 | | CRYSTAL STRUCTURE OF ASGPR 1 IN COMPLEX WITH GN-A. | | Descriptor: | 5-[(2~{R},3~{R},4~{R},5~{R},6~{R})-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-~{N}-[3-(propanoylamino)propyl]pentanamide, Asialoglycoprotein receptor 1, CALCIUM ION | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
6YG9
 
 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN (HSA) IN COMPLEX WITH GN-07. | | Descriptor: | 20-[[(2~{S})-5-[2-[2-[2-[2-[2-[2-(diethylamino)-2-oxidanylidene-ethoxy]ethoxy]ethylamino]-2-oxidanylidene-ethoxy]ethoxy]ethylamino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]amino]-20-oxidanylidene-icosanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-27 | | Release date: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
3QDH
 
 | |
3SNZ
 
 | |
3SO0
 
 | |
3SNY
 
 | |
3SO1
 
 | |
5XI1
 
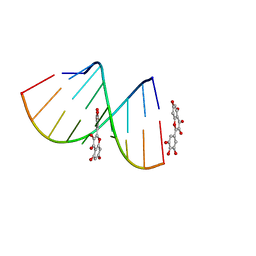 | | Structural Insight of Flavonoids binding to CAG repeat RNA that causes Huntington's Disease (HD) and Spinocerebellar Ataxia (SCAs) | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, RNA (5'-R(P*CP*CP*GP*CP*AP*GP*CP*GP*G)-3') | | Authors: | Tawani, A, Mishra, S.K, Khan, E, Kumar, A. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Myricetin Reduces Toxic Level of CAG Repeats RNA in Huntington's Disease (HD) and Spino Cerebellar Ataxia (SCAs).
ACS Chem. Biol., 13, 2018
|
|
6HHO
 
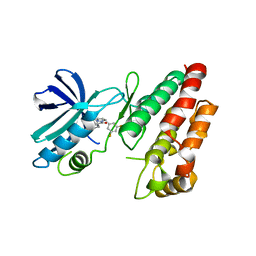 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
6MZ3
 
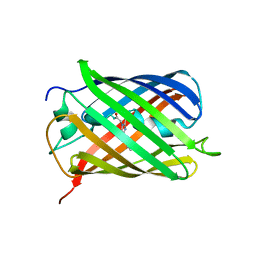 | | mCherry pH sensitive mutant - M66T (mCherryTYG) | | Descriptor: | PAmCherry1 protein | | Authors: | Haynes, E.P, Tantama, M. | | Deposit date: | 2018-11-03 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.088 Å) | | Cite: | Quantifying Acute Fuel and Respiration Dependent pH Homeostasis in Live Cells Using the mCherryTYG Mutant as a Fluorescence Lifetime Sensor.
Anal.Chem., 91, 2019
|
|
2WD5
 
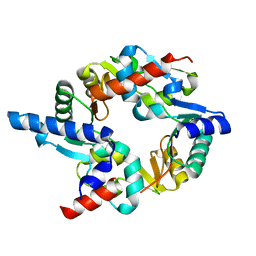 | | SMC hinge heterodimer (Mouse) | | Descriptor: | STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 1A, STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 3 | | Authors: | Michie, K.A, Haering, C.H, Nasmyth, K, Lowe, J. | | Deposit date: | 2009-03-20 | | Release date: | 2010-08-11 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Channel within the Smc1/Smc3 Hinge Required for Sister Chromatid Cohesion.
Embo J., 30, 2011
|
|
3TVY
 
 | |
3TW0
 
 | |
3TXA
 
 | | Structural Analysis of Adhesive Tip pilin, GBS104 from Group B Streptococcus agalactiae | | Descriptor: | CADMIUM ION, Cell wall surface anchor family protein, LITHIUM ION, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2011-09-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K4F
 
 | | Co-crystal structure of TNKS1 with compound 18 [4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide] | | Descriptor: | 4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X, Bregman, H, Wilson, C, DiMauro, E, Gunaydin, H. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4K4E
 
 | | Co-crystal structure of tnks1 with compound 52 [N~2-(5-chloro-2-methoxyphenyl)-N-[trans-4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)cyclohexyl]glycinamide] | | Descriptor: | N~2~-(5-chloro-2-methoxyphenyl)-N-[trans-4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)cyclohexyl]glycinamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MSK
 
 | | Co-crystal structure of tankyrase 1 with compound 34 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[4-(5-phenyl-1,3,4-oxadiazol-2-yl)phenyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MT9
 
 | | Co-crystal structure of tankyrase 1 with compound 49 | | Descriptor: | N-[trans-4-(4-cyanophenoxy)cyclohexyl]-3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MSG
 
 | | Crystal structure of tankyrase 1 with compound 22 | | Descriptor: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
3ENT
 
 | |
