3UYR
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, H-2 class I histocompatibility antigen, L-D alpha chain, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-06 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3UO1
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-11-16 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3V4U
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
7KGK
 
 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb16, Sybody-16, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
1B88
 
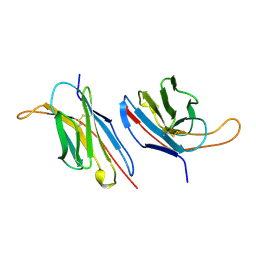 | | V-ALPHA 2.6 MOUSE T CELL RECEPTOR (TCR) DOMAIN | | Descriptor: | T CELL RECEPTOR V-ALPHA DOMAIN | | Authors: | Plaksin, D, Chacko, S, Navaza, J, Margulies, D.H, Padlan, E.A. | | Deposit date: | 1999-02-09 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray crystal structure of a Valpha2.6Jalpha38 mouse T cell receptor domain at 2.5 A resolution: alternate modes of dimerization and crystal packing.
J.Mol.Biol., 289, 1999
|
|
4G59
 
 | |
2MIZ
 
 | |
2O5N
 
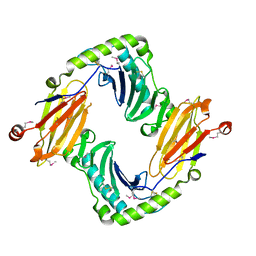 | | Crystal structure of a Viral Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuHV1gpm153, ... | | Authors: | Mans, J, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular Expression and Crystal Structure of the Murine Cytomegalovirus Major Histocompatibility Complex Class I-like Glycoprotein, m153.
J.Biol.Chem., 282, 2007
|
|
3E6F
 
 | | MHC CLASS I H-2Dd Heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PA9, from the Human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | BETA-2 MICROGLOBULIN, Envelope glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3ECB
 
 | | Crystal structure of mouse H-2Dd in complex with peptide P18-I10 derived from human immunodeficiency virus envelope glycoprotein 120 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2 microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Wang, R, Margulies, D.H. | | Deposit date: | 2008-08-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position
J.Immunol., 183, 2009
|
|
3E6H
 
 | | MHC CLASS I H-2Dd heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PI10, from the human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | Envelope glycoprotein 10-residue peptide, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3DMM
 
 | | Crystal structure of the CD8 alpha beta/H-2Dd complex | | Descriptor: | Beta-2 microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Margulies, D.H. | | Deposit date: | 2008-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position.
J.Immunol., 183, 2009
|
|
1U58
 
 | | Crystal structure of the murine cytomegalovirus MHC-I homolog m144 | | Descriptor: | MHC-I homolog m144, beta-2-microglobulin | | Authors: | Natarajan, K, Hicks, A, Robinson, H, Guan, R, Margulies, D.H. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
J.Mol.Biol., 358, 2006
|
|
3V52
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, ... | | Authors: | Mage, M.G, Dolan, M.A, Wang, R, Boyd, L.F, Revilleza, M.J, Robinson, H, Natarajan, K, Myers, N.B, Hansen, T.H, Margulies, D.H. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ8
 
 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ9
 
 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | Descriptor: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQA
 
 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
7TUD
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 6mer EEFGRC and dipeptide GL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, EEFGRC peptide, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUG
 
 | | Crystal structure of Tapasin in complex with PaSta2-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUH
 
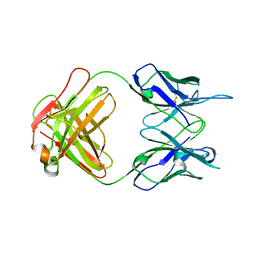 | | Crystal structure of anti-tapasin PaSta2-Fab | | Descriptor: | PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUE
 
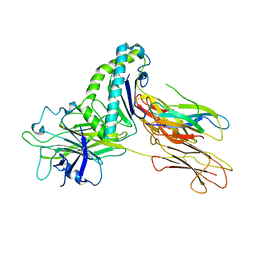 | | Crystal structure of Tapasin in complex with HLA-B*44:05 (T73C) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUF
 
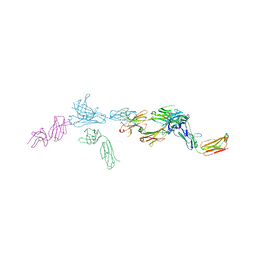 | | Crystal structure of Tapasin in complex with PaSta1-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta1 Fab heavy chain, PaSta1 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUC
 
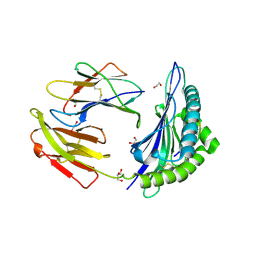 | | Crystal structure of HLA-B*44:05 (T73C) with 9mer EEFGRAFSF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
5WES
 
 | | Crystal Structure H2-Dd with disulfide-linked 5mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
