9N94
 
 | |
9NCQ
 
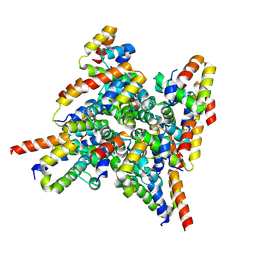 | | Cryo-EM structure of Fas-FADD complex | | Descriptor: | FAS-associated death domain protein, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Fosuah, E, Lin, Q, Shen, Z, Fu, T.M. | | Deposit date: | 2025-02-17 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Assembly and activation of the death-inducing signaling complex.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6DZP
 
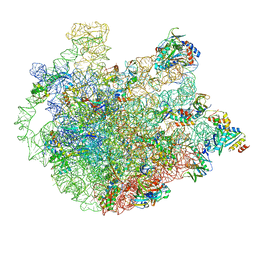 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DZK
 
 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 30S ribosomal subunit with MPY | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DZI
 
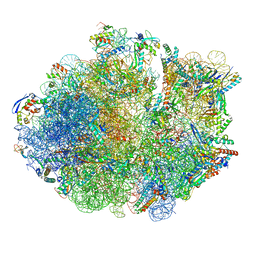 | | Cryo-EM Structure of Mycobacterium smegmatis 70S C(minus) ribosome 70S-MPY complex | | Descriptor: | 16S rRNA, 23 S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8Y2R
 
 | | NMR Solution Structure of the 2:1 Coptisine-ATF4-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*TP*AP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*AP*GP*GP*GP*AP*GP*C*(LIG)*(LIG))-3') | | Authors: | Xiao, C.M, Li, Y.P, Liu, Y.S, Dong, R.F, He, X.Y, Lin, Q, Zang, X, Wang, K.B, Xia, Y.Z, Kong, L.Y. | | Deposit date: | 2024-01-27 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | SOLUTION NMR | | Cite: | Overcoming Cancer Persister Cells by Stabilizing the ATF4 Promoter G-quadruplex.
Adv Sci, 11, 2024
|
|
4G35
 
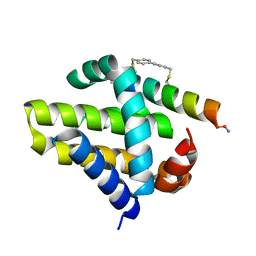 | | Mcl-1 in complex with a biphenyl cross-linked Noxa peptide. | | Descriptor: | 4,4'-bis(bromomethyl)biphenyl, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa BH3 peptide (cysteine-mediated cross-linked) | | Authors: | Drake, E, Edwardraja, S, Lin, Q, Gulick, A.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-12-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
2PY2
 
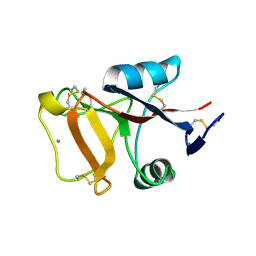 | | Structure of Herring Type II Antifreeze Protein | | Descriptor: | Antifreeze protein type II, CALCIUM ION | | Authors: | Liu, Y, Li, Z, Lin, Q, Seetharaman, J, Sivaraman, J, Hew, C.-L. | | Deposit date: | 2007-05-15 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Evolutionary Origin of Ca-Dependent Herring Type II Antifreeze Protein.
PLoS ONE, 2, 2007
|
|
4R05
 
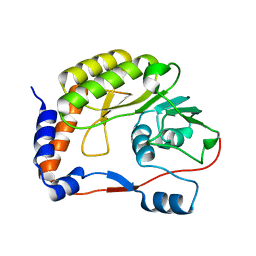 | | Crystal structure of the refolded DENV3 methyltransferase | | Descriptor: | Nonstructural protein NS5 | | Authors: | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
8JMZ
 
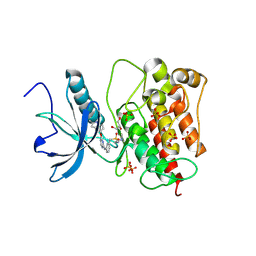 | | FGFR1 kinase domain with sulfatinib | | Descriptor: | Fibroblast growth factor receptor 1, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
8JOT
 
 | |
6A0Z
 
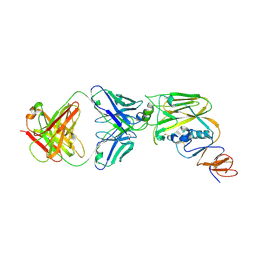 | | Crystal structure of broadly neutralizing antibody 13D4 bound to H5N1 influenza hemagglutinin, HA head region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 13D4, Fab Heavy Chain, ... | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
6A0X
 
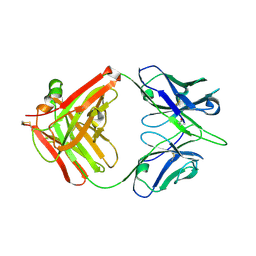 | | Crystal structure of broadly neutralizing antibody 13D4 | | Descriptor: | Antibody 13D4, Fab Heavy Chain, Fab Light Chain | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
7WCL
 
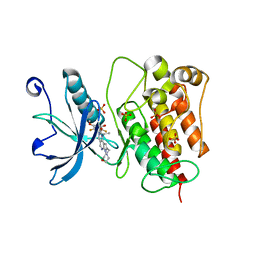 | | Crystal structure of FGFR1 kinase domain with Pemigatinib | | Descriptor: | 11-[2,6-bis(fluoranyl)-3,5-dimethoxy-phenyl]-13-ethyl-4-(morpholin-4-ylmethyl)-5,7,11,13-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),2(6),3,7-tetraen-12-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Lin, Q.M, Jiang, L.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants.
Commun Chem, 5, 2022
|
|
7LZ3
 
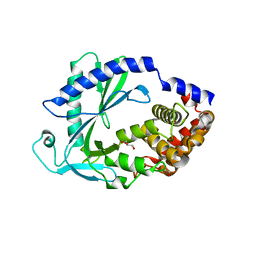 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7KXS
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9U7S
 
 | | FGFR2 kinase domain with a macrocyclic compound 8r | | Descriptor: | (E)-4-methyl-17-(1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl)-7,10-dioxa-4-aza-1(3,6)-imidazo[1,2-b]pyridazina-2(1,3)-benzenacyclodecaphan-3-one, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2025-03-25 | | Release date: | 2026-01-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of the First Novel Macrocycle-Based FGFR Inhibitors That Overcome Clinically Acquired Resistance.
J.Med.Chem., 2026
|
|
9U7E
 
 | | FGFR2 kinase domain with a macrocyclic compound 8g | | Descriptor: | (E)-4-methyl-17-(1-methyl-1H-pyrazol-4-yl)-7,10-dioxa-4-aza-1(3,6)-imidazo[1,2-b]pyridazina-2(3,5)-pyridinacyclodecaphan-3-one, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2025-03-24 | | Release date: | 2026-01-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of the First Novel Macrocycle-Based FGFR Inhibitors That Overcome Clinically Acquired Resistance.
J.Med.Chem., 2026
|
|
8KH8
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH6
 
 | | Crystal structure of FGFR4 kinase domain with 8r | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[5-bromanyl-6-(hydroxymethyl)-3-methoxy-pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH9
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH7
 
 | | Crystal structure of FGFR4 kinase domain with 8zc | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
9IVB
 
 | |
9PUK
 
 | |
9PUO
 
 | | Neutralizing monoclonal antibody Fab fragment bound to leptin | | Descriptor: | Leptin, Neutralizing antibody Fab fragment, heavy chain, ... | | Authors: | Tomchick, D.R, Wynn, R.M, Scherer, P.E. | | Deposit date: | 2025-07-31 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Leptin as a key driver for organ fibrogenesis.
Sci Adv, 11, 2025
|
|
