4ERS
 
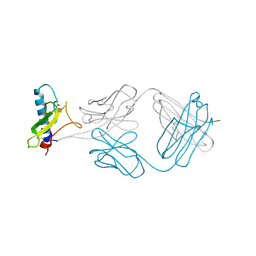 | | A Molecular Basis for Negative Regulation of the Glucagon Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Murray, J.M, Koth, C.M, Mukund, S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Molecular basis for negative regulation of the glucagon receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HV2
 
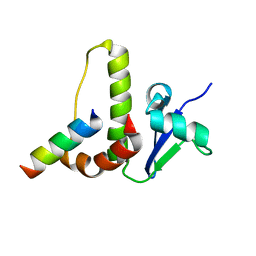 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
6N4R
 
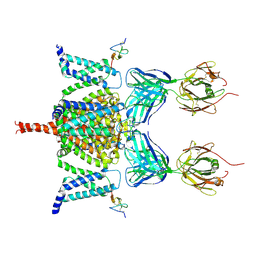 | | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
5EK0
 
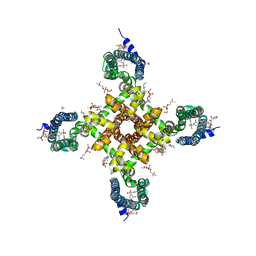 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
6N4Q
 
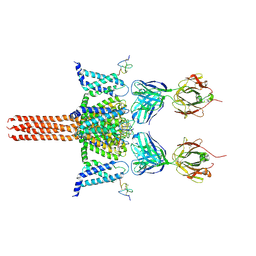 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
6N4I
 
 | | Structural basis of Nav1.7 inhibition by a gating-modifier spider toxin | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Nav1.7 VSD2-NavAb channel chimera protein, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Xu, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.541 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
6BPP
 
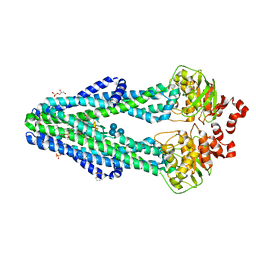 | | E. coli MsbA in complex with LPS and inhibitor G092 | | Descriptor: | (2E)-3-{6-[(1S)-1-(3-amino-2,6-dichlorophenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
6BPL
 
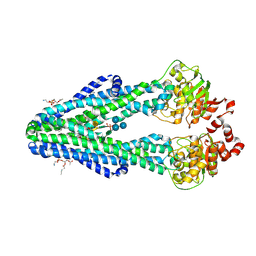 | | E. coli MsbA in complex with LPS and inhibitor G907 | | Descriptor: | (2E)-3-{6-[(1S)-1-(2-chloro-6-cyclopropylphenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-amino-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
2BBJ
 
 | | Crystal structure of the CorA Mg2+ transporter | | Descriptor: | divalent cation transport-related protein | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Zhang, R, Joachimiak, A, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter
Nature, 440, 2006
|
|
2BBH
 
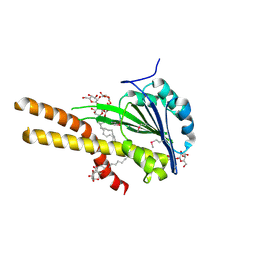 | | X-ray structure of T.maritima CorA soluble domain | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter.
Nature, 440, 2006
|
|
7SEL
 
 | | E. coli MsbA in complex with LPS and inhibitor G7090 (compound 3) | | Descriptor: | (2E)-3-{7-[(1S)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-1-methylnaphthalen-2-yl}prop-2-enoic acid, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Payandeh, J, Koth, C.M, Verma, V.A. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.978 Å) | | Cite: | Discovery of Inhibitors of the Lipopolysaccharide Transporter MsbA: From a Screening Hit to Potent Wild-Type Gram-Negative Activity.
J.Med.Chem., 65, 2022
|
|
3N7S
 
 | | Crystal structure of the ectodomain complex of the CGRP receptor, a Class-B GPCR, reveals the site of drug antagonism | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, N-{(1S)-5-amino-1-[(4-pyridin-4-ylpiperazin-1-yl)carbonyl]pentyl}-3,5-dibromo-Nalpha-{[4-(2-oxo-1,4-dihydroquinazolin-3 (2H)-yl)piperidin-1-yl]carbonyl}-D-tyrosinamide, N~4~-(5-cyclopropyl-1H-pyrazol-3-yl)-N~2~-1H-indazol-5-yl-6-methylpyrimidine-2,4-diamine, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-15 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ectodomain Complex of the CGRP Receptor, a Class-B GPCR, Reveals the Site of Drug Antagonism.
Structure, 18, 2010
|
|
3N7P
 
 | |
3N7R
 
 | | Crystal structure of the ectodomain complex of the CGRP receptor, a Class-B GPCR, reveals the site of drug antagonism | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide, N-{(1S)-5-amino-1-[(4-pyridin-4-ylpiperazin-1-yl)carbonyl]pentyl}-3,5-dibromo-Nalpha-{[4-(2-oxo-1,4-dihydroquinazolin-3 (2H)-yl)piperidin-1-yl]carbonyl}-D-tyrosinamide, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-15 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Ectodomain Complex of the CGRP Receptor, a Class-B GPCR, Reveals the Site of Drug Antagonism.
Structure, 18, 2010
|
|
6VJA
 
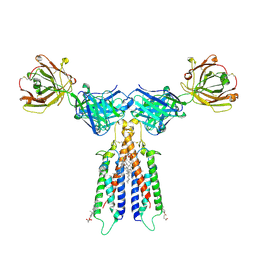 | | Structure of CD20 in complex with rituximab Fab | | Descriptor: | B-lymphocyte antigen CD20, CHOLESTEROL HEMISUCCINATE, Rituximab Fab heavy chain, ... | | Authors: | Rohou, A, Croll, T.I. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab.
Science, 367, 2020
|
|
7SI1
 
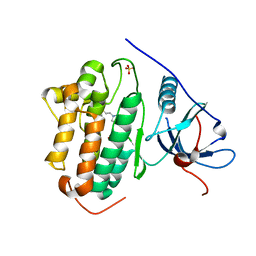 | |
7SHV
 
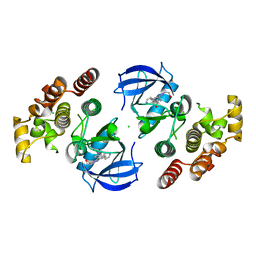 | | Crystal structure of BRAF kinase domain bound to GDC0879 | | Descriptor: | 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Kung, J.E, Sudhamsu, J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Clearing the Path to Rapid High-Quality Protein Purification
To Be Published
|
|
4LEX
 
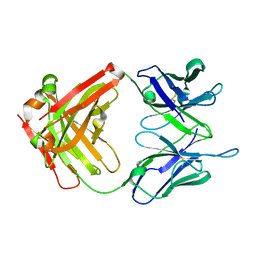 | | Unliganded crystal structure of mAb7 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Murray, J.M, Mukund, S. | | Deposit date: | 2013-06-26 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor.
J.Biol.Chem., 288, 2013
|
|
4LF3
 
 | |
4LY1
 
 | |
4LXZ
 
 | | Structure of Human HDAC2 in complex with SAHA (vorinostat) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone Deacetylase (HDAC) Inhibitor Kinetic Rate Constants Correlate with Cellular Histone Acetylation but Not Transcription and Cell Viability.
J.Biol.Chem., 288, 2013
|
|
4NNO
 
 | |
4NNP
 
 | |
