3KD8
 
 | |
3SOZ
 
 | | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2 | | Descriptor: | Cytoplasmic Protein STM1381, GLYCEROL | | Authors: | Joachimiak, A, Duke, N.E.C, Jedrzejczak, R, Li, H, Adkins, J, Brown, R, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2
To be Published
|
|
2QMM
 
 | |
3LM7
 
 | | Crystal Structure of DUF1341 representative, from Yersinia enterocolitica subsp. enterocolitica 8081 | | Descriptor: | BROMIDE ION, POTASSIUM ION, putative 4-Hydroxy-2-oxoglutarate aldolase / 2-dehydro-3-deoxyphosphogluconate aldolase | | Authors: | Joachimiak, A, Duke, N.E.C, Feldmann, B, Wu, R, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of DUF1341 representative, from Yersinia enterocolitica subsp. enterocolitica 8081
To be Published
|
|
3IDD
 
 | |
3OT6
 
 | | Crystal Structure of an enoyl-CoA hydratase/isomerase family protein from Psudomonas syringae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Joachimiak, A, Duke, N.E.C, Stein, A, Chhor, G, Freeman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-10 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an enoyl-CoA hydratase/isomerase family protein from Psudomonas syringae
To be Published
|
|
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
3OVK
 
 | |
3PN9
 
 | |
3R0A
 
 | |
5ERE
 
 | | Extracellular ligand binding receptor from Desulfohalobium retbaense DSM5692 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXO-4-METHYLPENTANOIC ACID, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Cuff, M, Wu, R, Endres, M, Pokkuluri, P.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-14 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel extracellular ligand receptor
To Be Published
|
|
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | Descriptor: | GROEL (HSP60 CLASS) | | Authors: | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1995-03-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
9EWK
 
 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | Descriptor: | Crambin, ETHANOL | | Authors: | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | Deposit date: | 2024-04-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
6VWW
 
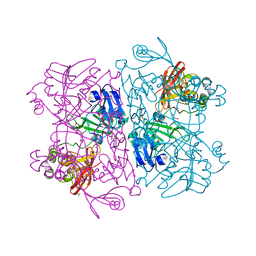 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
9D5W
 
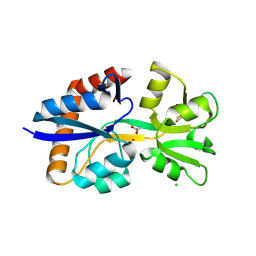 | | Crystal Structure of the Substrate Binding Domain Protein of the ABC Transporter PBP2_YxeM from Vibrio cholerae | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding portion, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Substrate Binding Domain Protein of the ABC Transporter PBP2_YxeM from Vibrio cholerae
To Be Published
|
|
9DCG
 
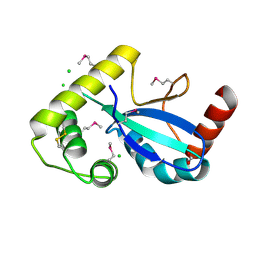 | | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-26 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae
To Be Published
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1Q8B
 
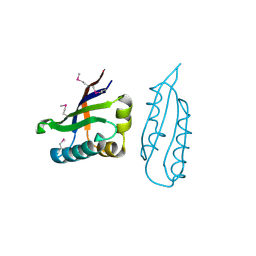 | |
1R0U
 
 | | Crystal structure of ywiB protein from Bacillus subtilis | | Descriptor: | GLYCEROL, protein ywiB | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of ywiB protein from Bacillus subtilis
to be published
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | Authors: | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-05-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
4XXT
 
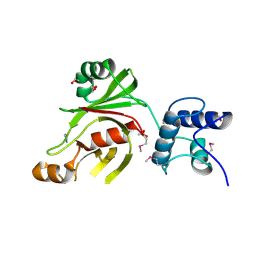 | | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fusion of predicted Zn-dependent amidase/peptidase (Cell wall hydrolase/DD-carboxypeptidase family) and uncharacterized domain of ErfK family peptodoglycan-binding domain, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from from Clostridium acetobutylicum ATCC 824
To Be Published
|
|
2AO9
 
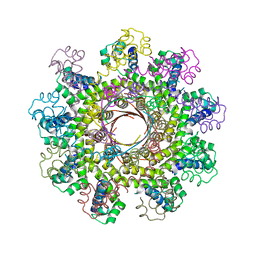 | |
7S6O
 
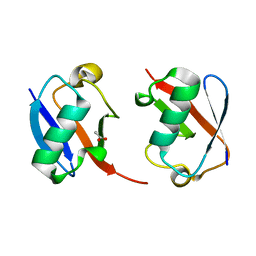 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
7S6P
 
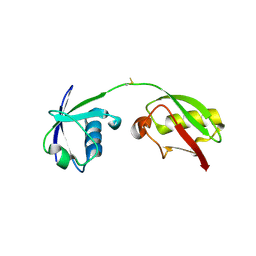 | | The crystal structure of human ISG15 | | Descriptor: | Ubiquitin-like protein ISG15 | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
