9J26
 
 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | Triple-helix region of human collagen type IV | | Authors: | Fan, X, Zhai, Y, Chu, Y, Fu, S, Li, D, Cao, K, Feng, P, Wu, X, Cai, H, Ma, L, Qian, S. | | Deposit date: | 2024-08-06 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
9J1T
 
 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | GLYCEROL, Triple-helix region of human collagen type IV | | Authors: | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Feng, P, Cao, K, Wu, X, Cai, H, Wang, H, Qian, S. | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
9J1R
 
 | | Structure of a triple-helix region of human Collagen type II from Trautec | | Descriptor: | SULFATE ION, Triple-helix region of human collagen type II | | Authors: | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Cao, K, Feng, P, Wang, X, Le, H, Tang, D, Zhang, F, Qian, S. | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
9J23
 
 | | Structure of a triple-helix region of human Collagen type II from Trautec | | Descriptor: | Triple-helix region of human collagen type II | | Authors: | Fan, X, Zhai, Y, Chu, Y, Fu, S, Li, D, Feng, P, Cao, K, Li, J, Si, Y, Ma, L, Qian, S. | | Deposit date: | 2024-08-06 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
4QWQ
 
 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | Descriptor: | Response regulator SaeR | | Authors: | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | Deposit date: | 2014-07-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8K6Z
 
 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
8FHD
 
 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6M3W
 
 | | Post-fusion structure of SARS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Fan, X, Cao, D, Zhang, X. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.
Nat Commun, 11, 2020
|
|
6J6K
 
 | | Apo-state streptavidin | | Descriptor: | Streptavidin | | Authors: | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
6J6J
 
 | | Biotin-bound streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
6LOZ
 
 | | crystal structure of alpha-momorcharin in complex with adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LP0
 
 | | crystal structure of alpha-momorcharin in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.519 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOR
 
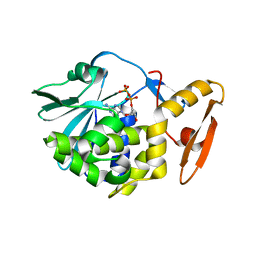 | | crystal structure of alpha-momorcharin in complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOW
 
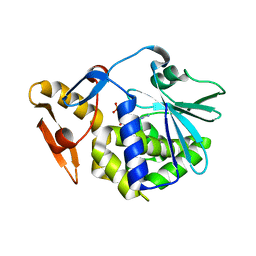 | | crystal structure of alpha-momorcharin in complex with GMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOV
 
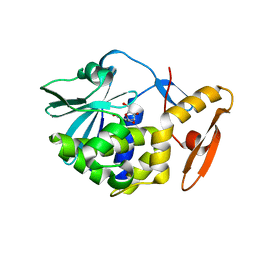 | | crystal structure of alpha-momorcharin in complex with xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOQ
 
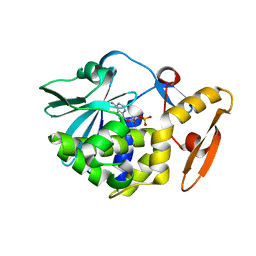 | | crystal structure of alpha-momorcharin in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOY
 
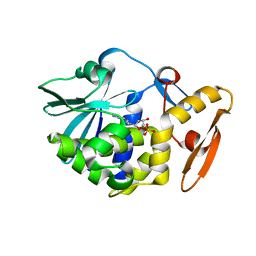 | | crystal structure of alpha-momorcharin in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
8IIC
 
 | |
8IIB
 
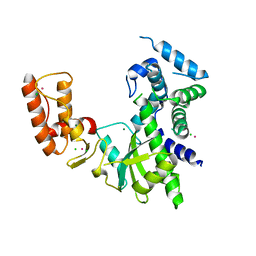 | | Crystal structure of Israeli acute paralysis virus RNA-dependent RNA polymerase delta85 mutant (residues 86-546) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fang, X, Lu, G, Hou, C, Gong, P. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Virol Sin, 38, 2023
|
|
3KM8
 
 | |
8S9B
 
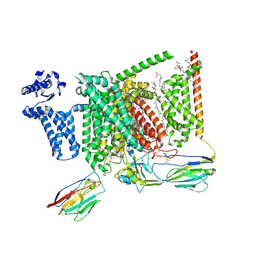 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S9C
 
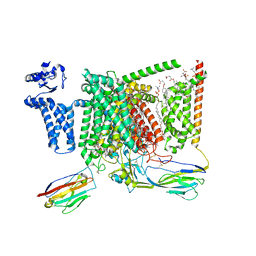 | | Cryo-EM structure of Nav1.7 with CBZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8THH
 
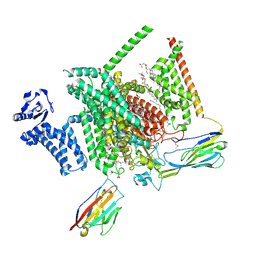 | | Cryo-EM structure of Nav1.7 with LTG | | Descriptor: | (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8THG
 
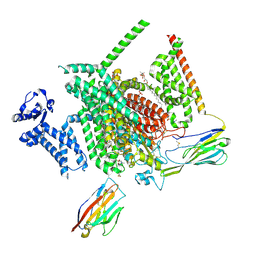 | | Cryo-EM structure of Nav1.7 with RLZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5DDZ
 
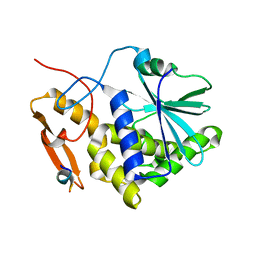 | | Crystal structure of the RTA-c10-P2 complex | | Descriptor: | 60S acidic ribosomal protein P2, Ricin | | Authors: | Zhu, Y, Fan, X, Wang, C, Niu, L, Li, X, Teng, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin
Sci Rep, 6, 2016
|
|
