2ABW
 
 | | Glutaminase subunit of the plasmodial PLP synthase (Vitamin B6 biosynthesis) | | Descriptor: | Pdx2 protein, TETRAETHYLENE GLYCOL | | Authors: | Gengenbacher, M, Fitzpatrick, T.B, Raschle, T, Flicker, K, Sinning, I, Mueller, S, Macheroux, P, Tews, I, Kappes, B. | | Deposit date: | 2005-07-17 | | Release date: | 2006-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Vitamin B6 Biosynthesis by the Malaria Parasite Plasmodium falciparum: Biochemical and structural insights
J.Biol.Chem., 281, 2006
|
|
2AJB
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-methyl-L-valyl-L-prolyl-L-isoleucine, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
9GD3
 
 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
9GD1
 
 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
9GD2
 
 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
9GD0
 
 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
1ORW
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1ORV
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2VP8
 
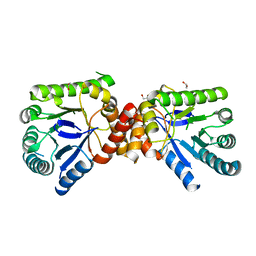 | | Structure of Mycobacterium tuberculosis Rv1207 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPTEROATE SYNTHASE 2 | | Authors: | Gengenbacher, M, Xu, T, Niyomwattanakit, P, Spraggon, G, Dick, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biochemical and Structural Characterization of the Putative Dihydropteroate Synthase Ortholog Rv1207 of Mycobacterium Tuberculosis.
Fems Microbiol.Lett., 287, 2008
|
|
2AJD
 
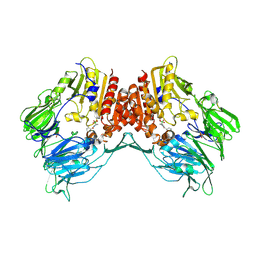 | | Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) | | Descriptor: | (2R)-N-[(2R)-2-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-N-[(5R)-5-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-L-PROLINAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
2AJC
 
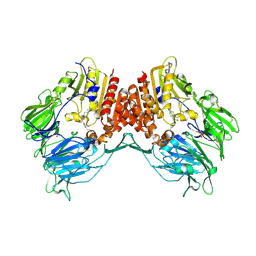 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
2AJ8
 
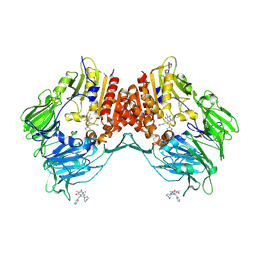 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
4XRM
 
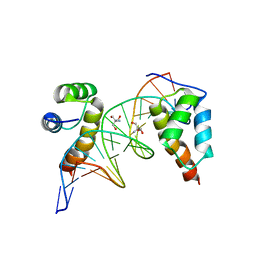 | | homodimer of TALE type homeobox transcription factor MEIS1 complexes with specific DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*AP*GP*CP*TP*GP*AP*CP*AP*GP*CP*TP*GP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*CP*AP*GP*CP*TP*GP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Morgunova, E, Jorma, A, Yin, Y, Nitta, K.R, Dave, K, Enge, M, Kivioja, T, Popov, A, Taipale, J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
4XRS
 
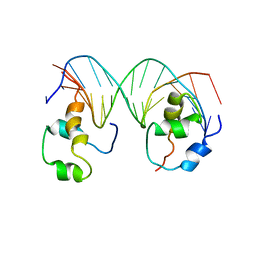 | | Heterodimeric complex of transcription factors MEIS1 and DLX3 on specific DNA | | Descriptor: | DNA (5'-D(P*AP*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*GP*AP*TP*AP*AP*TP*TP*GP*TP*T)-3'), ... | | Authors: | Jorma, A, Yin, Y, Nitta, K.R, Dave, K, Enge, M, Kivioja, T, Popov, A, Morgunova, E, Taipale, J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
5BNG
 
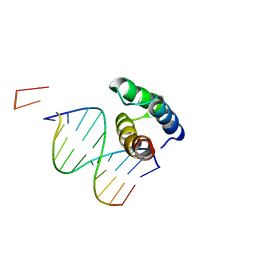 | | monomer of TALE type homeobox transcription factor MEIS1 complexes with specific DNA | | Descriptor: | DNA (5'-D(P*AP*AP*TP*TP*AP*GP*CP*TP*GP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A-3'), ... | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Nitta, K, Dave, K, Popov, A, Taipale, M, Enge, M, Kivioja, T, Taipale, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
7NKY
 
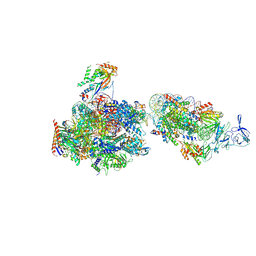 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NKX
 
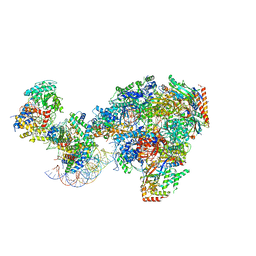 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3H25
 
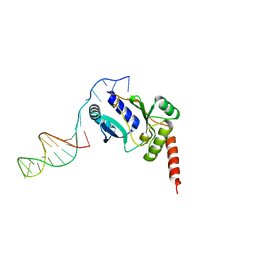 | | Crystal structure of the catalytic domain of primase Repb' in complex with initiator DNA | | Descriptor: | Replication protein B, SINGLE STRANDED INITIATOR DNA (SSIA) | | Authors: | Geibel, S, Banchenko, S, Engel, M, Lanka, E, Saenger, W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H20
 
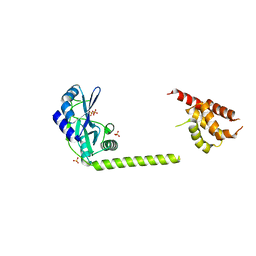 | | Crystal structure of primase RepB' | | Descriptor: | DIPHOSPHATE, Replication protein B, SULFATE ION | | Authors: | Geibel, S, Banchenko, S, Engel, M, Lanka, E, Saenger, W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6FVG
 
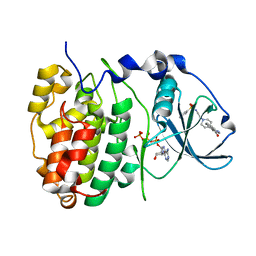 | | The Structure of CK2alpha with CCh507 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6FVF
 
 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
2F93
 
 | | K Intermediate Structure of Sensory Rhodopsin II/Transducer Complex in Combination with the Ground State Structure | | Descriptor: | RETINAL, Sensory rhodopsin II, Sensory rhodopsin II transducer, ... | | Authors: | Moukhametzianov, R.I, Klare, J.P, Efremov, R.G, Baecken, C, Goeppner, A, Labahn, J, Engelhard, M, Bueldt, G, Gordeliy, V.I. | | Deposit date: | 2005-12-05 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of the signal in sensory rhodopsin and its transfer to the cognate transducer.
Nature, 440, 2006
|
|
2F95
 
 | | M intermediate structure of sensory rhodopsin II/transducer complex in combination with the ground state structure | | Descriptor: | RETINAL, Sensory rhodopsin II, Sensory rhodopsin II transducer, ... | | Authors: | Moukhametzianov, R.I, Klare, J.P, Efremov, R.G, Baecken, C, Goeppner, A, Labahn, J, Engelhard, M, Bueldt, G, Gordeliy, V.I. | | Deposit date: | 2005-12-05 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of the signal in sensory rhodopsin and its transfer to the cognate transducer.
Nature, 440, 2006
|
|
4GYC
 
 | | Structure of the SRII(D75N mutant)/HtrII Complex in I212121 space group ("U" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2012-09-05 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0501 Å) | | Cite: | Ground state structure of D75N mutant of sensory rhodopsin II in complex with its cognate transducer.
J Photochem Photobiol B, 123C, 2013
|
|
5JJF
 
 | | Structure of the SRII/HtrII Complex in I212121 space group ("U" shape) - M state | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-23 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
