8X16
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF101 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures of adenosine receptor A3AR bound to selective agonists
Nat Commun, 15, 2024
|
|
8X17
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF102 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of adenosine receptor A3AR bound to selective agonists
Nat Commun, 15, 2024
|
|
8HDO
 
 | | Structure of A2BR bound to synthetic agonists BAY 60-6583 | | Descriptor: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Adenosine A2b receptor, CHOLESTEROL, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structures of adenosine receptor A 2B R bound to endogenous and synthetic agonists.
Cell Discov, 8, 2022
|
|
8HDP
 
 | | Structure of A2BR bound to endogenous agonists adenosine | | Descriptor: | ADENOSINE, Adenosine A2b receptor, CHOLESTEROL, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of adenosine receptor A 2B R bound to endogenous and synthetic agonists.
Cell Discov, 8, 2022
|
|
7CZ0
 
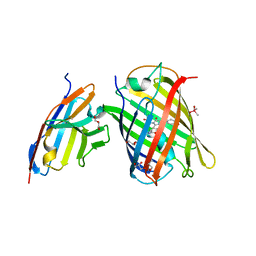 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb92) | | Descriptor: | ACETATE ION, CACODYLATE ION, CACODYLIC ACID, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-09-06 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | An improved fluorescent protein tag and its nanobodies for membrane protein expression, stability assay, and purification
To Be Published
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
1C3E
 
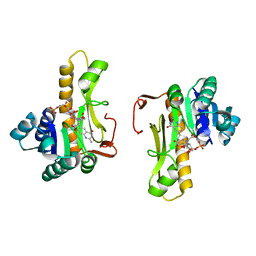 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1C2T
 
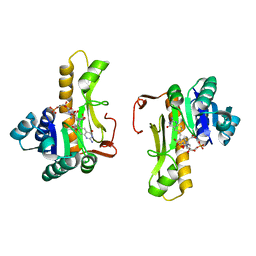 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1JKX
 
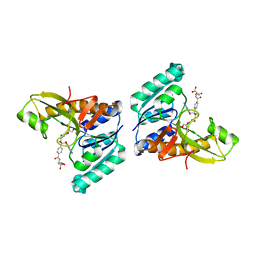 | | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase | | Descriptor: | N-[5'-O-PHOSPHONO-RIBOFURANOSYL]-2-[2-HYDROXY-2-[4-[GLUTAMIC ACID]-N-CARBONYLPHENYL]-3-[2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL]-PROPANYLAMINO]-ACETAMIDE, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Greasley, S.E, Marsilje, T.H, Cai, H, Baker, S, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase.
Biochemistry, 40, 2001
|
|
8H0Q
 
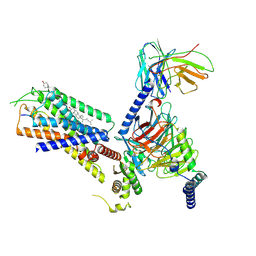 | | Structure of the GRP14-27-GRPR-Gq complex | | Descriptor: | CHOLESTEROL, G-alpha q, GRP, ... | | Authors: | Li, C, Xu, Y, Liu, H, Cai, H, Xu, H.E, Yin, W. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of itch-associated neuropeptides by bombesin receptors
Cell Res., 33, 2023
|
|
8H0P
 
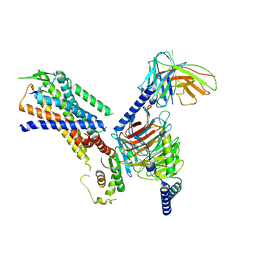 | | Structure of the NMB30-NMBR and Gq complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Xu, Y, Liu, H, Cai, H, Xu, H.E, Yin, W. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular recognition of itch-associated neuropeptides by bombesin receptors
Cell Res., 33, 2023
|
|
7CYV
 
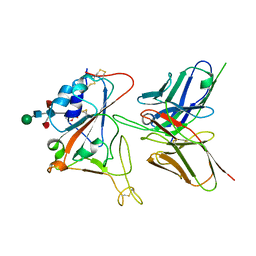 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
3BJ1
 
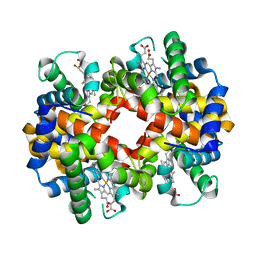 | | met-Perch Hemoglobin at pH 5.7 | | Descriptor: | ACETYL GROUP, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha, ... | | Authors: | Aranda IV, R, Cai, H, Levin, E.J, Richards, M.P, Phillips Jr, G.N. | | Deposit date: | 2007-12-02 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of fish versus mammalian hemoglobins: Effect of the heme pocket environment on autooxidation and hemin loss.
Proteins, 75, 2008
|
|
3BJ2
 
 | | met-Perch Hemoglobin at pH 6.3 | | Descriptor: | ACETYL GROUP, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha, ... | | Authors: | Aranda IV, R, Cai, H, Levin, E.J, Richards, M.P, Phillips Jr, G.N. | | Deposit date: | 2007-12-02 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of fish versus mammalian hemoglobins: Effect of the heme pocket environment on autooxidation and hemin loss.
Proteins, 75, 2008
|
|
3BJ3
 
 | | met-Perch hemoglobin at pH 8.0 | | Descriptor: | ACETYL GROUP, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha, ... | | Authors: | Aranda IV, R, Cai, H, Levin, E.J, Richards, M.P, Phillips Jr, G.N. | | Deposit date: | 2007-12-02 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of fish versus mammalian hemoglobins: Effect of the heme pocket environment on autooxidation and hemin loss.
Proteins, 75, 2008
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7D2Z
 
 | | Structure of sybody SR31 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7D30
 
 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7WKD
 
 | | TRH-TRHR G protein complex | | Descriptor: | Gq, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Y, Cai, H, You, C, He, X, Yuan, Q, Jiang, H, Cheng, X, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into ligand binding and activation of the human thyrotropin-releasing hormone receptor.
Cell Res., 32, 2022
|
|
7U5B
 
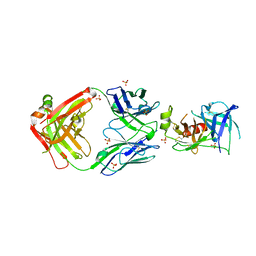 | | Structure of Human KLK5 bound to anti-KLK5 Fab | | Descriptor: | Kallikrein-5, SULFATE ION, anti-KLK5 Fab Heavy Chain, ... | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Dual antibody inhibition of KLK5 and KLK7 for Netherton syndrome and atopic dermatitis.
Sci Transl Med, 14, 2022
|
|
7MWY
 
 | |
7MWZ
 
 | |
7V6Z
 
 | | Cryo-EM structure of Patched1 (V1084A mutant) in lipid nanodisc, 3.64 angstrom (reprocessed with the dataset of 7dzp) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
