5D7X
 
 | |
7O2X
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound T180 | | Descriptor: | ACETATE ION, N-[(6-methylimidazo[1,2-a]pyridin-2-yl)methyl]-4-oxidanylidene-pyrido[1,2-a]pyrimidine-2-carboxamide, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex with compound T180
To Be Published
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4TWN
 
 | | Human EphA3 Kinase domain in complex with Birb796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
6FGT
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
8BS4
 
 | |
8BS6
 
 | |
8BS5
 
 | |
7ACD
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound T30 (UZH1a) | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-oxidanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes.
Chemmedchem, 16, 2021
|
|
7BC2
 
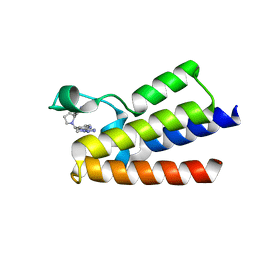 | | BAZ2A bromodomain in complex with triazole compound MS04-TN04 | | Descriptor: | 2-(1-methyl-1H-1,2,3-triazol-5-yl)-5-((2-(pyridin-4-yl)pyrrolidin-1-yl)methyl)-1H-benzo[d]imidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
6EPV
 
 | | The ATAD2 bromodomain in complex with compound 5 | | Descriptor: | (2~{R})-2-azanyl-~{N}-(4-oxidanylidene-6,7-dihydro-5~{H}-1,3-benzothiazol-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPX
 
 | | The ATAD2 bromodomain in complex with compound 3 | | Descriptor: | (2~{R})-2-carbamimidamido-~{N}-(4-ethanoyl-1,3-thiazolidin-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPU
 
 | | The ATAD2 bromodomain in complex with compound 2 | | Descriptor: | (2~{S})-~{N}-(4-ethanoyl-1,3-thiazol-2-yl)piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
5H85
 
 | |
9QIU
 
 | | Crystal structure of YTHDF2 in complex with compound 13 (AI-DF2-56) | | Descriptor: | 3-sulfanyl-1,2,4-triazin-5-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-17 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9QEM
 
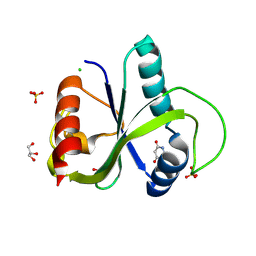 | | Crystal structure of YTHDF2 in complex with compound 3 (AI-DF2-11) | | Descriptor: | 5-[[(3-methoxyphenyl)amino]methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-10 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9QEO
 
 | |
9QFL
 
 | | Crystal structure of YTHDF2 in complex with compound 15 (AI-DF2-68) | | Descriptor: | 6-sulfanylidene-1,7-dihydropyrazolo[3,4-d]pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-11 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9QEL
 
 | |
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
6YM2
 
 | | Crystal structure of YTHDC1 with compound ADO_AC_25 | | Descriptor: | SULFATE ION, YTHDC1, ~{N},9-dimethylpurin-6-amine | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of YTHDC1 with compound ADO_AC_25
To Be Published
|
|
7B7G
 
 | | BAZ2A bromodomain in complex with compounds MS04 and B11 | | Descriptor: | 1-[1-(3,5-dimethoxyphenyl)piperidin-4-yl]-2,3-dimethyl-guanidine, 5-ethyl-2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7B82
 
 | | BAZ2A bromodomain in complex with triazole compound MS04-TN03 | | Descriptor: | (2~{S})-1-[4-[[(~{E})-~{N},~{N}'-dimethylcarbamimidoyl]amino]phenyl]-~{N}-[[2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazol-5-yl]methyl]pyrrolidine-2-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7B7I
 
 | | BAZ2A bromodomain in complex with triazole compound MS04-TN02 | | Descriptor: | 2-[4-[[(~{E})-~{N},~{N}'-dimethylcarbamimidoyl]amino]piperidin-1-yl]-~{N}-[[2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazol-5-yl]methyl]ethanamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
