2MFZ
 
 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
6NRV
 
 | |
4UMV
 
 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN THE E2P STATE | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
4UMW
 
 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN E2.PI STATE | | Descriptor: | MAGNESIUM ION, TETRAFLUOROALUMINATE ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
7W7R
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7W7S
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
9EVC
 
 | |
9EUQ
 
 | | CryoEM structure of LMCA1 in E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase lmo0841, MAGNESIUM ION | | Authors: | Prabudiansyah, I, Andersson, M. | | Deposit date: | 2024-03-28 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Dephosphorylation and ion binding in prokaryotic calcium transport.
Sci Adv, 10, 2024
|
|
6ETJ
 
 | | HUMAN PFKFB3 IN COMPLEX WITH KAN0438241 | | Descriptor: | 4-[[3-(5-fluoranyl-2-oxidanyl-phenyl)phenyl]sulfonylamino]-2-oxidanyl-benzoic acid, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Gustafsson, N.M.S, Lundback, T, Farnegardh, K, Groth, P, Wiitta, E, Jonsson, M, Hallberg, K, Pennisi, R, Huguet Ninou, A, Martinsson, J, Norstrom, C, Schultz, J, Andersson, M, Markova, N, Marttila, P, Norin, M, Olin, T, Helleday, T. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun, 9, 2018
|
|
6F22
 
 | | Complex between MTH1 and compound 29 (a 4-amino-2,7-diazaindole derivative) | | Descriptor: | (3~{S})-3-phenyl-4-(2~{H}-pyrazolo[3,4-b]pyridin-4-yl)morpholine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F1X
 
 | | Complex between MTH1 and compound 7 (a 7-azaindole-2-amide derivative) | | Descriptor: | 4-(3-chlorophenyl)-~{N}-ethyl-1~{H}-pyrrolo[2,3-b]pyridine-2-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F23
 
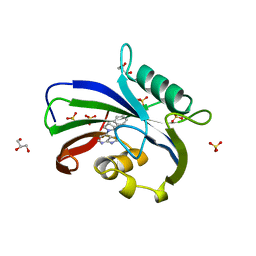 | | Complex between MTH1 and compound 16 (a 4-amino-7-azaindole derivative) | | Descriptor: | 4-[(2~{R})-2-phenylpyrrolidin-1-yl]-1~{H}-pyrrolo[2,3-b]pyridine, 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, ... | | Authors: | Viklund, J, Tresaugues, L, Talagas, A, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
7Z6N
 
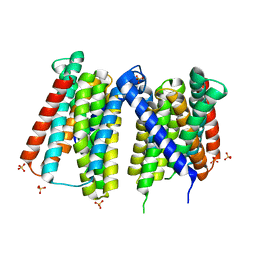 | | Crystal structure of Zn2+-transporter BbZIP in a metal-stripped state | | Descriptor: | Putative membrane protein, SULFATE ION | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
7Z6M
 
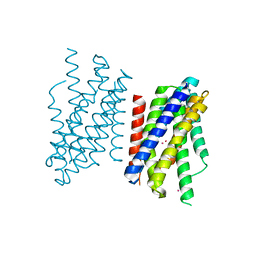 | | Crystal structure of Zn2+-transporter BbZIP in a cadmium bound state | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
6F20
 
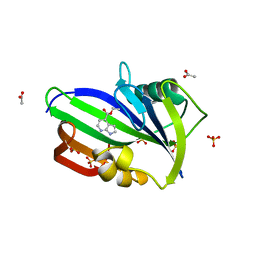 | | Complex between MTH1 and compound 1 (a 7-azaindole-4-ester derivative) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, Ethyl 1H-pyrrolo[2,3-b]pyridine-4-carboxylate, ... | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6FMD
 
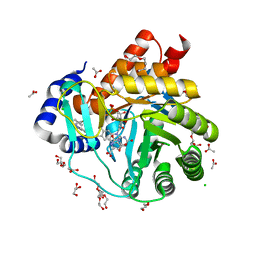 | | Targeting myeloid differentiation using potent human dihydroorotate dehydrogenase (hDHODH) inhibitors based on 2-hydroxypyrazolo[1,5-a]pyridine scaffold | | Descriptor: | 2-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Jarva, M, Andersson, M, Lolli, M.L, Friemann, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting Myeloid Differentiation Using Potent 2-Hydroxypyrazolo[1,5- a]pyridine Scaffold-Based Human Dihydroorotate Dehydrogenase Inhibitors.
J. Med. Chem., 61, 2018
|
|
5MVC
 
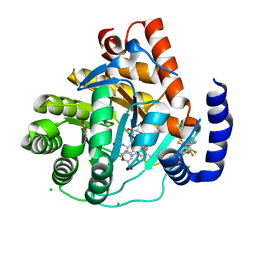 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 4-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,5-thiadiazole-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
5NHY
 
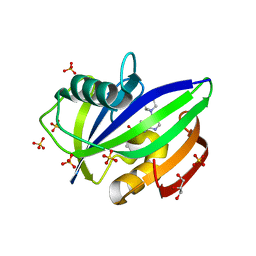 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
5MVD
 
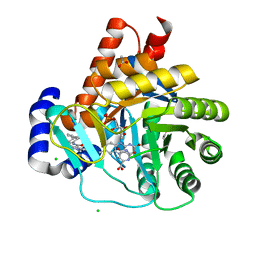 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 1,5-dimethyl-3-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
5MUT
 
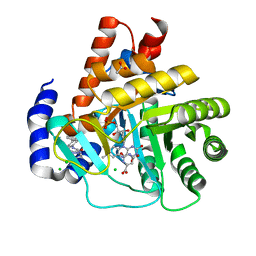 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
2X5U
 
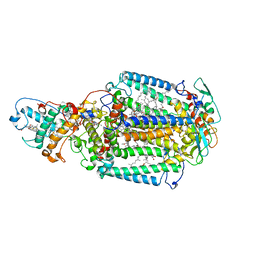 | | 80 microsecond Laue diffraction snapshot from crystals of a photosynthetic reaction centre without illumination. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-induced structural changes in a photosynthetic reaction center caught by Laue diffraction.
Science, 328, 2010
|
|
2X5V
 
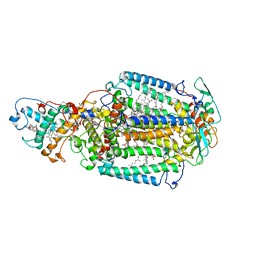 | | 80 microsecond laue diffraction snapshot from crystals of a photosynthetic reaction centre 3 millisecond following photoactivation. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction.
Science, 328, 2010
|
|
1IGV
 
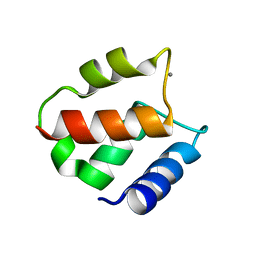 | | BOVINE CALBINDIN D9K BINDING MN2+ | | Descriptor: | MANGANESE (II) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IG5
 
 | | BOVINE CALBINDIN D9K BINDING MG2+ | | Descriptor: | MAGNESIUM ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M, Svensson, L.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
