1KBF
 
 | | Solution Structure of the Cysteine-Rich C1 Domain of Kinase Suppressor of Ras | | Descriptor: | Kinase suppressor of Ras 1, ZINC ION | | Authors: | Zhou, M, Horita, D.A, Waugh, D.S, Byrd, R.A, Morrison, D.K. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).
J.Mol.Biol., 315, 2002
|
|
3JXE
 
 | | Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Yang, B, Ding, J. | | Deposit date: | 2009-09-19 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of P. horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase
To be Published
|
|
6M4G
 
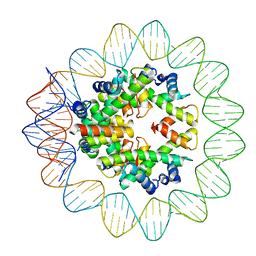 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6M4H
 
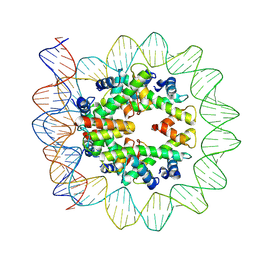 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (103-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6M4D
 
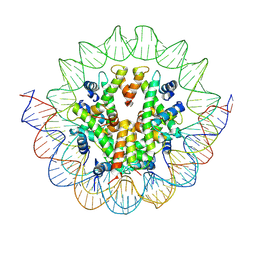 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (125-MER), Histone H2A.V, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
3PPY
 
 | | Crystal structure of the D1596A/N1602A double mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPW
 
 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
1VGK
 
 | | The crystal structure of class I Major histocompatibility complex, H-2Kd at 2.0 A resolution | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Zhou, M, Xu, Y, Liu, Y, Gao, G.F, Tien, P, Rao, Z. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The crystal structure of class I major histocompatibility complex, H-2Kd at 2.0 A resolution
To be Published
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
3KT6
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with Trp | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3KT0
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase. | | Descriptor: | (2S)-1-{[(2S)-3-(2-methoxyethoxy)-2-methylpropyl]oxy}propan-2-amine, Tryptophanyl-tRNA synthetase, cytoplasmic | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3KT8
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with L-tryptophanamide | | Descriptor: | L-TRYPTOPHANAMIDE, SULFATE ION, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3KT3
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
1KBE
 
 | | Solution structure of the cysteine-rich C1 domain of Kinase Suppressor of Ras | | Descriptor: | Kinase Suppressor of Ras, ZINC ION | | Authors: | Zhou, M, Horita, D.A, Waugh, D.S, Byrd, R.A, Morrison, D.K. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-23 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).
J.Mol.Biol., 315, 2002
|
|
5B63
 
 | | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg) | | Descriptor: | Arginine--tRNA ligase, tRNA-Arg | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg)
To Be Published
|
|
5YYM
 
 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate Arg | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
5YYN
 
 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate TRNA(Arg) | | Descriptor: | Arginine--tRNA ligase, TRNA | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
9AYM
 
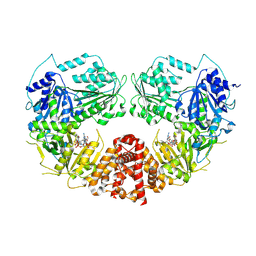 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-acetone-CoA bisubstrate probe | | Descriptor: | RNA cytidine acetyltransferase, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-[2-[3-[1-[(2~{R},3~{R},4~{R},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]sulfanyl-2-oxidanylidene-propyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9B0I
 
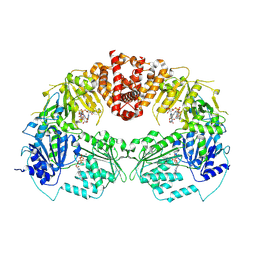 | |
9B0E
 
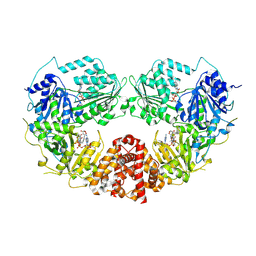 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-amide-CoA bisubstrate probe and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA cytidine acetyltransferase, [[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[[1-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]amino]-2-oxidanylidene-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
7UVE
 
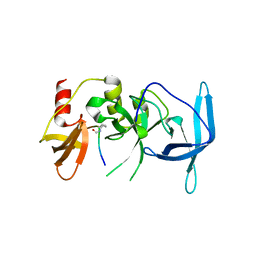 | | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac | | Descriptor: | Histone-lysine N-methyltransferase eggless, peptide H3K9me2K14ac | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac
To Be Published
|
|
7UW8
 
 | | Drosophila melanogaster setdb1-tuor domain | | Descriptor: | Histone-lysine N-methyltransferase eggless | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain
To Be Published
|
|
1J95
 
 | |
