139D
 
 | |
134D
 
 | |
1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
4IUR
 
 | |
6IEJ
 
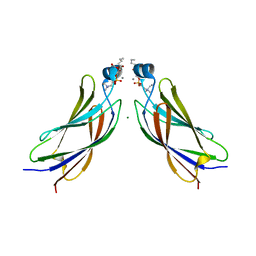 | | The C2 domain of cytosolic phospholipase A2 alpha bound to phosphatidylcholine | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CALCIUM ION, Cytosolic phospholipase A2, ... | | Authors: | Hirano, Y, Gao, Y.G, Stephenson, D.J, Vu, N.T, Malinina, L, Chalfant, C.E, Patel, D.J, Brown, R.E. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2alpha.
Elife, 8, 2019
|
|
3KHH
 
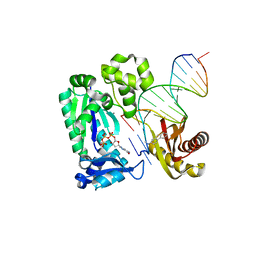 | | Dpo4 extension ternary complex with a C base opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-AMINOFLUORENE, 5'-D(*CP*CP*TP*A*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KHG
 
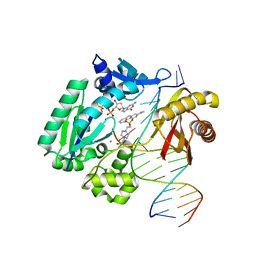 | | Dpo4 extension ternary complex with misinserted A opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-AMINOFLUORENE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KHL
 
 | | Dpo4 post-extension ternary complex with misinserted A opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7SDE
 
 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
7SJR
 
 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
3RAX
 
 | |
3RV1
 
 | | Crystal structure of the N-terminal and RNase III domains of K. polysporus Dcr1 E224Q mutant | | Descriptor: | K. polysporus Dcr1 | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RW6
 
 | | Structure of nuclear RNA export factor TAP bound to CTE RNA | | Descriptor: | Nuclear RNA export factor 1, constitutive transport element(CTE)of Mason-Pfizer monkey virus RNA | | Authors: | Teplova, M, Khin, N.W, Patel, D.J, Izaurralde, E. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RWV
 
 | | Crystal Structure of apo-form of Human Glycolipid Transfer Protein at 1.5 A resolution | | Descriptor: | Glycolipid transfer protein, SULFATE ION | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0K
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RW7
 
 | | Structure of N-terminal domain of nuclear RNA export factor TAP | | Descriptor: | Nuclear RNA export factor 1 | | Authors: | Teplova, M, Khin, N.W, Wohlbold, L, Izaurralde, E, Patel, D.J. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6PPR
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PPJ
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), IRON/SULFUR CLUSTER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2A5R
 
 | | Complex of tetra-(4-n-methylpyridyl) porphin with monomeric parallel-stranded DNA tetraplex, snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, C-MYC promoter, NMR, 6 struct. | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
2A5P
 
 | | Monomeric parallel-stranded DNA tetraplex with snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, NMR, 8 struct. | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
201D
 
 | |
202D
 
 | |
2ASL
 
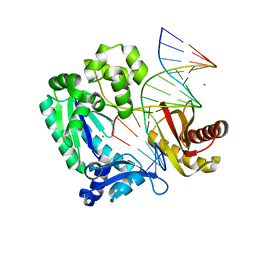 | | oxoG-modified Postinsertion Binary Complex | | Descriptor: | 5'-D(*CP*T*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DOC))-3', CALCIUM ION, ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-23 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
2ANR
 
 | | Crystal structure (II) of Nova-1 KH1/KH2 domain tandem with 25nt RNA hairpin | | Descriptor: | 5'-R(*CP*(5BU)P*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
