1MCP
 
 | | PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS | | Descriptor: | IGA-KAPPA MCPC603 FAB (HEAVY CHAIN), IGA-KAPPA MCPC603 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Satow, Y, Cohen, G.H, Padlan, E.A, Davies, D.R. | | Deposit date: | 1984-07-09 | | Release date: | 1985-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A.
J.Mol.Biol., 190, 1986
|
|
1UM2
 
 | |
1AT5
 
 | | HEN EGG WHITE LYSOZYME WITH A SUCCINIMIDE RESIDUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, LYSOZYME, ... | | Authors: | Noguchi, S, Miyawaki, K, Satow, Y. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Succinimide and isoaspartate residues in the crystal structures of hen egg-white lysozyme complexed with tri-N-acetylchitotriose.
J.Mol.Biol., 278, 1998
|
|
1EHL
 
 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
1AT6
 
 | | HEN EGG WHITE LYSOZYME WITH A ISOASPARTATE RESIDUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Noguchi, S, Miyawaki, K, Satow, Y. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Succinimide and isoaspartate residues in the crystal structures of hen egg-white lysozyme complexed with tri-N-acetylchitotriose.
J.Mol.Biol., 278, 1998
|
|
3VW3
 
 | | Antibody 64M-5 Fab in complex with a double-stranded DNA (6-4) photoproduct | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), COBALT HEXAMMINE(III), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y. | | Deposit date: | 2012-07-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a double-stranded DNA (6-4) photoproduct in complex with the 64M-5 antibody Fab
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ITQ
 
 | | HUMAN RENAL DIPEPTIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RENAL DIPEPTIDASE, ZINC ION | | Authors: | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | Deposit date: | 2002-02-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
1ITU
 
 | | HUMAN RENAL DIPEPTIDASE COMPLEXED WITH CILASTATIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CILASTATIN, RENAL DIPEPTIDASE, ... | | Authors: | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | Deposit date: | 2002-02-03 | | Release date: | 2002-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
3M7O
 
 | | Crystal structure of mouse MD-1 in complex with phosphatidylcholine | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Harada, H, Ohto, U, Satow, Y. | | Deposit date: | 2010-03-17 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse MD-1 with endogenous phospholipid bound in its cavity
J.Mol.Biol., 400, 2010
|
|
1KEG
 
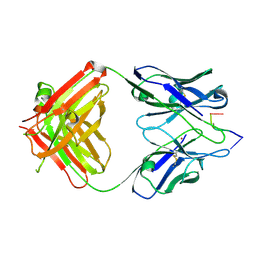 | | Antibody 64M-2 Fab complexed with dTT(6-4)TT | | Descriptor: | 5'-D(*TP*(64T)P*TP*T)-3', Anti-(6-4) photoproduct antibody 64M-2 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-2 Fab (light chain), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Sato, K, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DNA (6-4) photoproduct dTT(6-4)TT in complex with the 64M-2 antibody Fab fragment implies increased antibody-binding affinity by the flanking nucleotides.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1NGQ
 
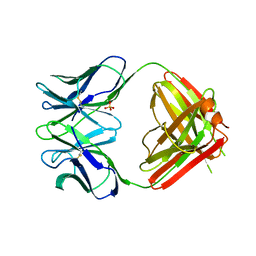 | | N1G9 (IGG1-LAMBDA) FAB FRAGMENT | | Descriptor: | N1G9 (IGG1-LAMBDA), SULFATE ION | | Authors: | Mizutani, R, Satow, Y. | | Deposit date: | 1995-06-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structures of the Fab fragment of murine N1G9 antibody from the primary immune response and of its complex with (4-hydroxy-3-nitrophenyl)acetate.
J.Mol.Biol., 254, 1995
|
|
1NGP
 
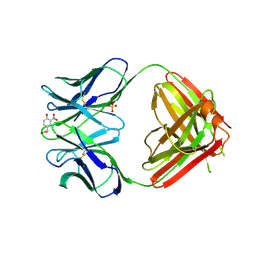 | |
1UJ3
 
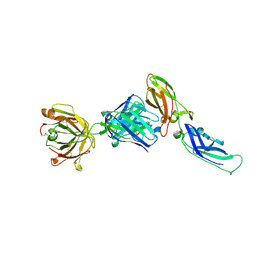 | | Crystal structure of a humanized Fab fragment of anti-tissue-factor antibody in complex with tissue factor | | Descriptor: | IgG Fab heavy chain, IgG Fab light chain, tissue factor | | Authors: | Ohto, U, Mizutani, R, Nakamura, M, Adachi, H, Satow, Y. | | Deposit date: | 2003-07-25 | | Release date: | 2004-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a humanized Fab fragment of anti-tissue-factor antibody in complex with tissue factor.
J.Synchrotron Radiat., 11, 2004
|
|
1A0F
 
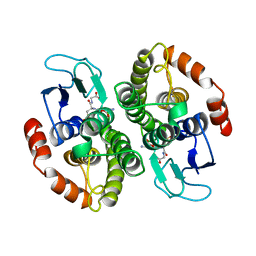 | | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTATHIONESULFONIC ACID | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID | | Authors: | Nishida, M, Harada, S, Noguchi, S, Inoue, H, Takahashi, K, Satow, Y. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106.
J.Mol.Biol., 281, 1998
|
|
2E56
 
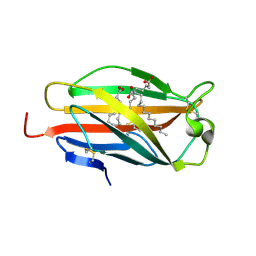 | | Crystal structure of human MD-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, MYRISTIC ACID | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2006-12-19 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa.
Science, 316, 2007
|
|
2E59
 
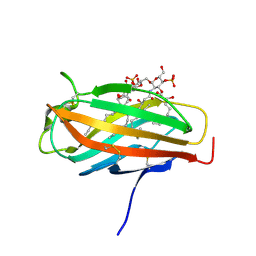 | | Crystal structure of human MD-2 in complex with lipid IVa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2006-12-19 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa
Science, 316, 2007
|
|
1JVA
 
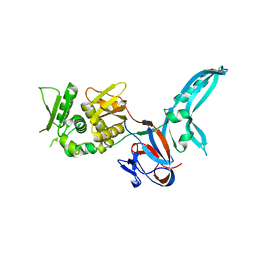 | |
1RTU
 
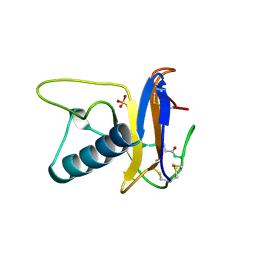 | | USTILAGO SPHAEROGENA RIBONUCLEASE U2 | | Descriptor: | RIBONUCLEASE U2, SULFATE ION | | Authors: | Noguchi, S, Satow, Y, Uchida, T, Sasaki, C, Matsuzaki, T. | | Deposit date: | 1995-05-12 | | Release date: | 1996-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
1RNH
 
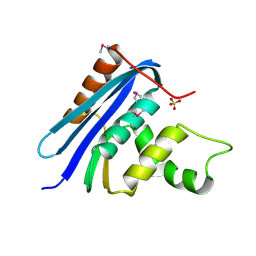 | | STRUCTURE OF RIBONUCLEASE H PHASED AT 2 ANGSTROMS RESOLUTION BY MAD ANALYSIS OF THE SELENOMETHIONYL PROTEIN | | Descriptor: | RIBONUCLEASE HI, SULFATE ION | | Authors: | Yang, W, Hendrickson, W.A, Crouch, R.J, Satow, Y. | | Deposit date: | 1990-07-11 | | Release date: | 1991-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein.
Science, 249, 1990
|
|
3AK8
 
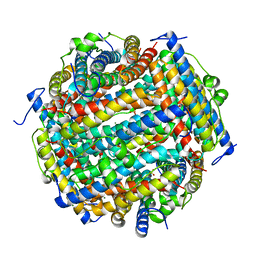 | | Crystal structure of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis | | Descriptor: | DNA protection during starvation protein, MAGNESIUM ION, SULFATE ION | | Authors: | Miyamoto, T, Asahina, Y, Miyazaki, S, Shimizu, H, Ohto, U, Noguchi, S, Satow, Y. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3AK9
 
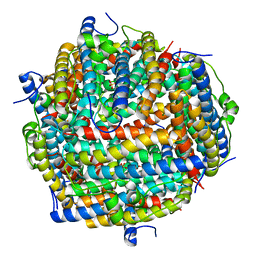 | | Crystal structure of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis, FE-soaked form | | Descriptor: | DNA protection during starvation protein, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Miyamoto, T, Asahina, Y, Miyazaki, S, Shimizu, H, Ohto, U, Noguchi, S, Satow, Y. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3W67
 
 | | Crystal structure of mouse alpha-tocopherol transfer protein in complex with alpha-tocopherol and phosphatidylinositol-(3,4)-bisphosphate | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Alpha-tocopherol transfer protein | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency
Science, 340, 2013
|
|
3W68
 
 | | Crystal structure of mouse alpha-tocopherol transfer protein in complex with alpha-tocopherol and phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, ... | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency
Science, 340, 2013
|
|
3AGX
 
 | |
3AGZ
 
 | |
