6COY
 
 | |
6COZ
 
 | |
6Q0J
 
 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
6Q0K
 
 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
6Q0T
 
 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
6NYB
 
 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Park, E, Rawson, S, Li, K, Jeon, H, Eck, M.J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
4R3R
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4R3P
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8DSW
 
 | |
5TR1
 
 | |
5TQQ
 
 | |
4V4N
 
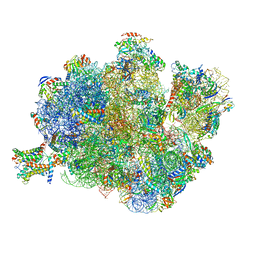 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
4G4E
 
 | | Crystal structure of the L88A mutant of HslV from Escherichia coli | | Descriptor: | ATP-dependent protease subunit HslV | | Authors: | Lee, J.W, Park, E, Yoo, H.M, Ha, B.H, An, J.Y, Jeon, Y.J, Seol, J.H, Eom, S.H, Chung, C.H. | | Deposit date: | 2012-07-16 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Structural Alteration in the Pore Motif of the Bacterial 20S Proteasome Homolog HslV Leads to Uncontrolled Protein Degradation
J.Mol.Biol., 425, 2013
|
|
8TQM
 
 | |
6D8E
 
 | |
6DUK
 
 | | EGFR with an allosteric inhibitor | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{1-oxo-6-[4-(piperazin-1-yl)phenyl]-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Park, E, Eck, M.J. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single and Dual Targeting of Mutant EGFR with an Allosteric Inhibitor.
Cancer Discov, 9, 2019
|
|
6XMT
 
 | | Structure of P5A-ATPase Spf1, BeF3-bound form | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMQ
 
 | | Structure of P5A-ATPase Spf1, AMP-PCP-bound form | | Descriptor: | MAGNESIUM ION, P5A-type ATPase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMS
 
 | | Structure of P5A-ATPase Spf1, AlF4-bound form | | Descriptor: | MAGNESIUM ION, P5A-type ATPase, TETRAFLUOROALUMINATE ION | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMU
 
 | |
6XMP
 
 | | Structure of P5A-ATPase Spf1, Apo form | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6PP9
 
 | | Crystal structure of BRAF:MEK1 complex | | Descriptor: | 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Park, E, Eck, M.J. | | Deposit date: | 2019-07-05 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
8DNY
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by decatransin | | Descriptor: | Decatransin peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNZ
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by apratoxin F | | Descriptor: | Apratoxin F peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNX
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by cotransin | | Descriptor: | Cotransin analogue peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
