5GOJ
 
 | | Lys63-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
3QQ3
 
 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | 9-mer peptide from Neuraminidase, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
3QQ4
 
 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, VP35 | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
3R93
 
 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
3RO3
 
 | | crystal structure of LGN/mInscuteable complex | | Descriptor: | CHLORIDE ION, ETHANOL, G-protein-signaling modulator 2, ... | | Authors: | Zhu, J, Wen, W, Shang, Y, Wei, Z, Pan, Z, Wang, W, Zhang, M. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
3RAF
 
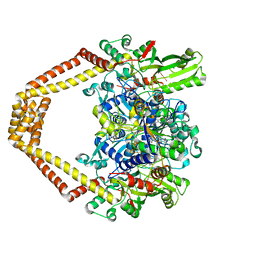 | | Quinazolinedione-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibitor-stabilised cleavage complexes of topoisomerase IIa: structural analysis of drug-dependent inter- and intramolecular interactions
To be Published
|
|
3RTP
 
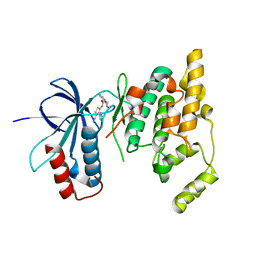 | | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration | | Descriptor: | Mitogen-activated protein kinase 10, N-[4-cyano-3-(1H-1,2,4-triazol-5-yl)thiophen-2-yl]-2-(2-oxo-3,4-dihydroquinolin-1(2H)-yl)acetamide | | Authors: | Bowers, S, Truong, A.P, Neitz, R.J, Hom, R.K, Sealy, J.M, Probst, G.D, Quincy, Q, Peterson, B, Chan, W, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Pan, H, Lin, M, Yao, N, Artis, D.R, Zhang, H, Chen, L, Dryer, M, Samant, B, Zmolek, W, Wong, K, Lorentzen, C, Goldbach, E, Tonn, G, Quinn, K.P, Sauer, J, Wright, S, Powell, K, Ruslim, L, Ren, Z, Bard, F, Yednock, T.A, Griswold-Prenne, I. | | Deposit date: | 2011-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5ITG
 
 | |
4HZU
 
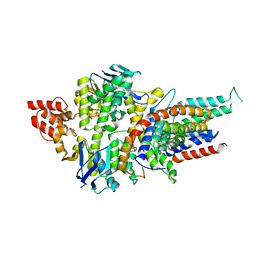 | | Structure of a bacterial energy-coupling factor transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA 1, Energy-coupling factor transporter ATP-binding protein EcfA 2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Wang, T.L, Fu, G.B, Pan, X.J, Shi, Y.G. | | Deposit date: | 2012-11-15 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structure of a bacterial energy-coupling factor transporter.
Nature, 497, 2013
|
|
4I3H
 
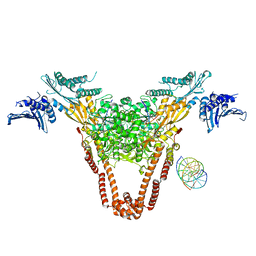 | | A three-gate structure of topoisomerase IV from Streptococcus pneumoniae | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*GP*CP*GP*GP*TP*AP*AP*TP*AP*CP*GP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*AP*AP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*TP*TP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*CP*GP*TP*AP*TP*TP*AP*CP*CP*GP*CP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Crevel, I, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2012-11-26 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.
Nucleic Acids Res., 41, 2013
|
|
3V6P
 
 | | Crystal structure of the DNA-binding domain of dHax3, a TAL effector | | Descriptor: | dHax3 | | Authors: | Deng, D, Yan, C.Y, Pan, X.J, Wang, J.W, Shi, Y.G, Yan, N. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Sequence-Specific Recognition of DNA by TAL Effectors
Science, 2012
|
|
1WUG
 
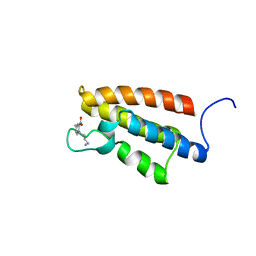 | | complex structure of PCAF bromodomain with small chemical ligand NP1 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-4-METHYL-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-07 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1WUM
 
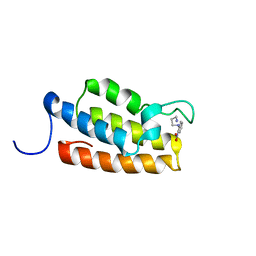 | | Complex structure of PCAF bromodomain with small chemical ligand NP2 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1XUT
 
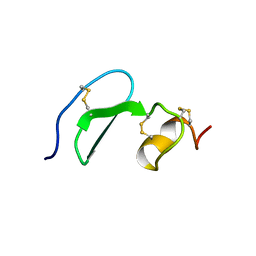 | | Solution structure of TACI-CRD2 | | Descriptor: | Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding.
J.Biol.Chem., 280, 2005
|
|
1XU1
 
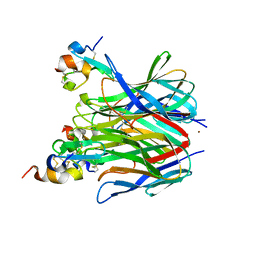 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1XUX
 
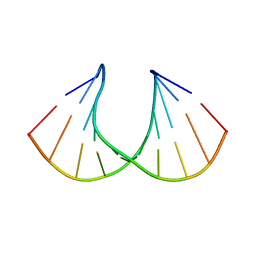 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMS)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
1XU2
 
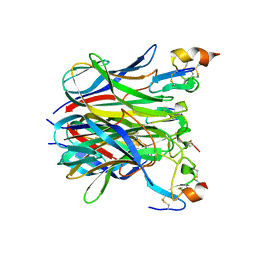 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1XT9
 
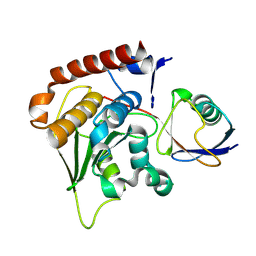 | | Crystal Structure of Den1 in complex with Nedd8 | | Descriptor: | Neddylin, Sentrin-specific protease 8 | | Authors: | Reverter, D, Wu, K, Erdene, T.G, Pan, Z.Q, Wilkinson, K.D, Lima, C.D. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Complex between Nedd8 and the Ulp/Senp Protease Family Member Den1.
J.Mol.Biol., 345, 2005
|
|
1XUW
 
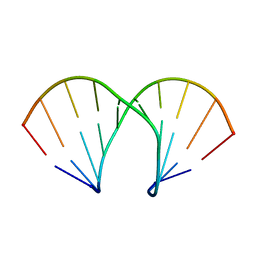 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMT)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
1YAU
 
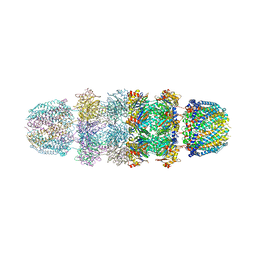 | | Structure of Archeabacterial 20S proteasome- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAR
 
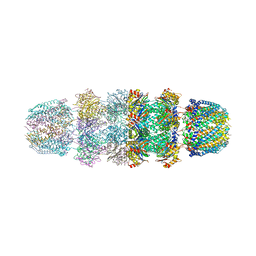 | | Structure of Archeabacterial 20S proteasome mutant D9S- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YU9
 
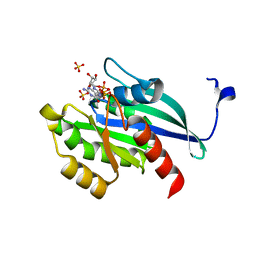 | | GppNHp-Bound Rab4A | | Descriptor: | GTP-binding protein, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
5D9B
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5CX3
 
 | | Crystal structure of FYCO1 LIR in complex with LC3A | | Descriptor: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Cheng, X, Pan, L. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
