6YRJ
 
 | | SFX structure of dye-type peroxidase DtpB in the ferric state | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Putative iron-dependent peroxidase | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7NES
 
 | |
7NER
 
 | |
7NET
 
 | |
6Z68
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | Acetyl esterase/lipase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
8GDU
 
 | | Crystal structure of a mutant methyl transferase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium (NESG) Target MvR53-11M | | Descriptor: | Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8GDY
 
 | | Crystal structure of the human PDI first domain with 9 mutations | | Descriptor: | 1,2-ETHANEDIOL, Protein disulfide-isomerase, THIOCYANATE ION | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
6OZ4
 
 | |
6Z69
 
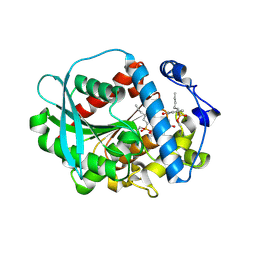 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6OZ2
 
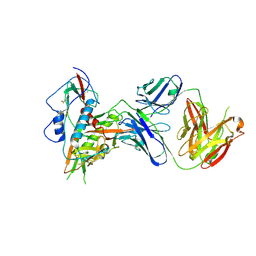 | |
8HDU
 
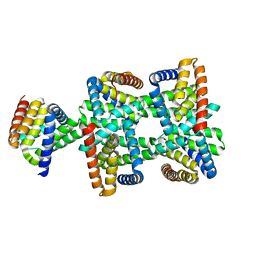 | |
8HDV
 
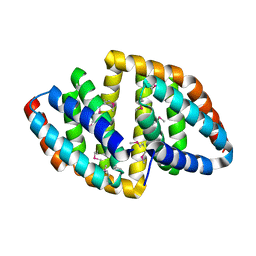 | |
8HFS
 
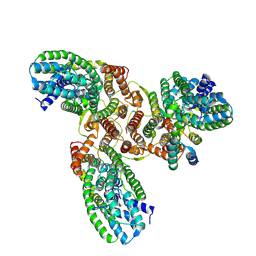 | | The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis | | Descriptor: | Bacteriocin lactococcin-A, Lactococcin-A immunity protein, Mannose-specific PTS system, ... | | Authors: | Wang, J.W. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-08 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural Basis of the Mechanisms of Action and Immunity of Lactococcin A, a Class IId Bacteriocin.
Appl.Environ.Microbiol., 89, 2023
|
|
7RCO
 
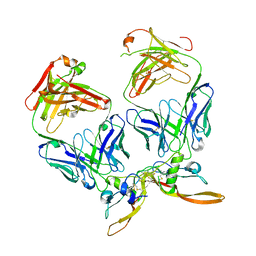 | |
6Q84
 
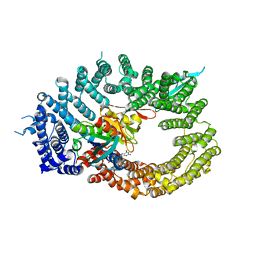 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
4UON
 
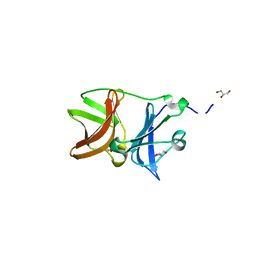 | |
7SAX
 
 | |
7SB2
 
 | |
7SAT
 
 | |
7SAZ
 
 | |
7T8W
 
 | | Structure of antibody 3G12 bound to Respiratory Syncytial Virus G central conserved domain mutant S177Q | | Descriptor: | 3G12 Fab Heavy chain, 3G12 Fab Light chain, Mature secreted glycoprotein G | | Authors: | Nunez Castrejon, A.M, O'Rourke, S.M, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design and Antigenic Validation of Respiratory Syncytial Virus G Immunogens.
J.Virol., 96, 2022
|
|
8RJC
 
 | |
8RJB
 
 | |
8RJD
 
 | |
