8UAC
 
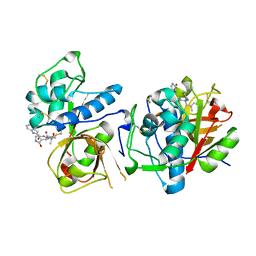 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CATHEPSIN L IN COMPLEX WITH AC1115
To be Published
|
|
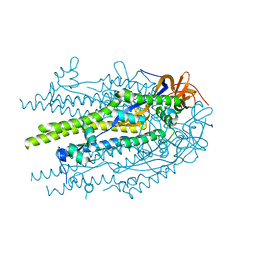
8U1G
 
 | |
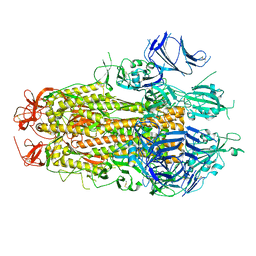
8TYL
 
 | |
8TYJ
 
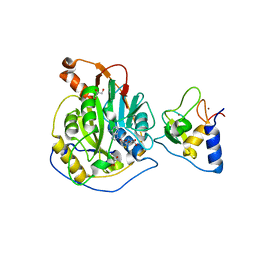 | |
8TYO
 
 | |
8Q93
 
 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | Descriptor: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | Authors: | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q95
 
 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | Descriptor: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | Authors: | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q94
 
 | |
8TV6
 
 | | SARS-CoV-2 Mac1 in complex with MDOLL-0169 | | Descriptor: | (1R,6R)-6-{[3-(methoxycarbonyl)-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophen-2-yl]carbamoyl}cyclohex-3-ene-1-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 2024
|
|
8TV7
 
 | | SARS-CoV-2 Mac1 in complex with MDOLL-0229 | | Descriptor: | (1R,2R)-2-{[3-(methoxycarbonyl)-4,5,6,7,8,9-hexahydrocycloocta[b]thiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, GLYCEROL, Papain-like protease nsp3 | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 2024
|
|
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q4O
 
 | |
8TOU
 
 | | ACE2-peptide 2 complex crystal form 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Franck, C, Payne, R.J, Christie, M. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOR
 
 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOT
 
 | | ACE2-peptide2 complex crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOQ
 
 | | ACE2-peptide 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOS
 
 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TMA
 
 | |
8TM1
 
 | |
8PYW
 
 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K3K
 
 | |
8TBE
 
 | | Co-crystal structure of SARS-CoV-2 Mpro with Pomotrelvir | | Descriptor: | 3C-like proteinase nsp5, Pomotrelvir bound form | | Authors: | Olland, A, Fontano, E, White, A. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evaluation of in vitro antiviral activity of SARS-CoV-2 M pro inhibitor pomotrelvir and cross-resistance to nirmatrelvir resistance substitutions.
Antimicrob.Agents Chemother., 67, 2023
|
|
