7BUS
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) | | Descriptor: | Chalcone synthase | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
5LQS
 
 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
2F6X
 
 | |
7CMC
 
 | | CRYSTAL STRUCTURE OF DEOXYHYPUSINE SYNTHASE FROM PYROCOCCUS HORIKOSHII | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable deoxyhypusine synthase | | Authors: | Yu, J, Gai, Z.Q, Okada, C, Yao, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexible NAD+Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A.
Int J Mol Sci, 21, 2020
|
|
6ZTU
 
 | |
6ZU3
 
 | |
7AZU
 
 | |
7XRQ
 
 | |
2JLC
 
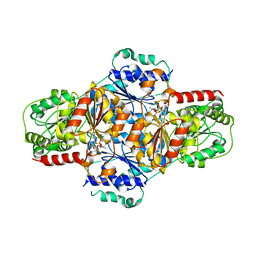 | | Crystal structure of E.coli MenD, 2-succinyl-5-enolpyruvyl-6-hydroxy- 3-cyclohexadiene-1-carboxylate synthase - native protein | | Descriptor: | 2-SUCCINYL-5-ENOLPYRUVYL-6-HYDROXY-3-CYCLOHEXENE -1-CARBOXYLATE SYNTHASE, MANGANESE (II) ION, THIAMINE DIPHOSPHATE | | Authors: | Dawson, A, Fyfe, P.K, Hunter, W.N. | | Deposit date: | 2008-09-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity and Reactivity in Menaquinone Biosynthesis: The Structure of Escherichia Coli Mend (2-Succinyl-5-Enolpyruvyl-6-Hydroxy-3-Cyclohexadiene-1-Carboxylate Synthase).
J.Mol.Biol., 384, 2008
|
|
1EA0
 
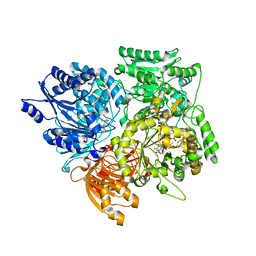 | | Alpha subunit of A. brasilense glutamate synthase | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Binda, C, Bossi, R.T, Vanoni, M.A, Mattevi, A. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-Talk and Ammonia Channeling between Active Centers in the Unexpected Domain Arrangement of Glutamate Synthase
Structure, 8, 2000
|
|
1FE2
 
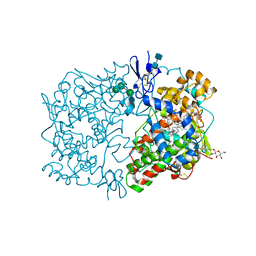 | | CRYSTAL STRUCTURE OF DIHOMO-GAMMA-LINOLEIC ACID BOUND IN THE CYCLOOXYGENASE CHANNEL OF PROSTAGLANDIN ENDOPEROXIDE H SYNTHASE-1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EICOSA-8,11,14-TRIENOIC ACID, PROSTAGLANDIN ENDOPEROXIDE H SYNTHASE-1, ... | | Authors: | Thuresson, E.D, Malkowski, M.G, Lakkides, K.M, Smith, W.L, Garavito, R.M. | | Deposit date: | 2000-07-20 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mutational and X-ray crystallographic analysis of the interaction of dihomo-gamma -linolenic acid with prostaglandin endoperoxide H synthases.
J.Biol.Chem., 276, 2001
|
|
7E38
 
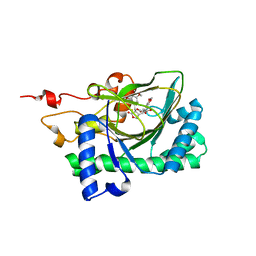 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with yatein and succinate | | Descriptor: | (3~{R},4~{R})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, (3~{S},4~{S})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, Deoxypodophyllotoxin synthase, ... | | Authors: | Wu, M.-H, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1IGX
 
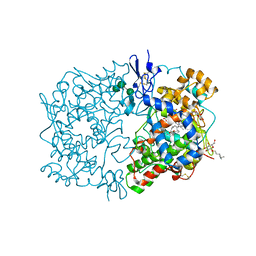 | | Crystal Structure of Eicosapentanoic Acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,8,11,14,17-EICOSAPENTAENOIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
1IGZ
 
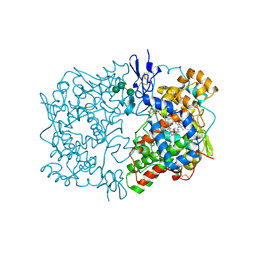 | | Crystal Structure of Linoleic acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
1WDW
 
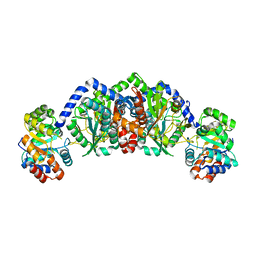 | | Structural basis of mutual activation of the tryptophan synthase a2b2 complex from a hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase alpha chain, Tryptophan synthase beta chain 1 | | Authors: | Lee, S.J, Ogasahara, K, Ma, J, Nishio, K, Ishida, M, Yamagata, Y, Tsukihara, T, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-19 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational Changes in the Tryptophan Synthase from a Hyperthermophile upon alpha(2)beta(2) Complex Formation: Crystal Structure of the Complex
Biochemistry, 44, 2005
|
|
3A5Q
 
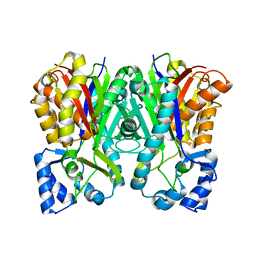 | | Benzalacetone synthase from Rheum palmatum | | Descriptor: | Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5S
 
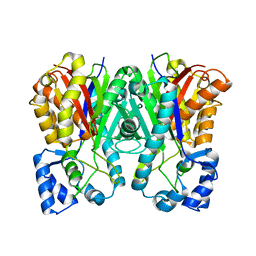 | | Benzalacetone synthase (I207L/L208F) | | Descriptor: | Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1I7Q
 
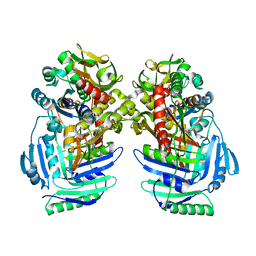 | | ANTHRANILATE SYNTHASE FROM S. MARCESCENS | | Descriptor: | ANTHRANILATE SYNTHASE, BENZOIC ACID, GLUTAMIC ACID, ... | | Authors: | Spraggon, G, Kim, C, Nguyen-Huu, X, Yee, M.-C, Yanofsky, C, Mills, S.E. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1I7S
 
 | | ANTHRANILATE SYNTHASE FROM SERRATIA MARCESCENS IN COMPLEX WITH ITS END PRODUCT INHIBITOR L-TRYPTOPHAN | | Descriptor: | ANTHRANILATE SYNTHASE, TRPG, TRYPTOPHAN | | Authors: | Spraggon, G, Kim, C, Nguyen-Huu, X, Yee, M.-C, Yanofsky, C, Mills, S.E. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2A59
 
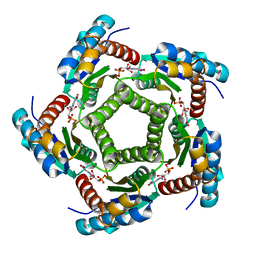 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
1XC4
 
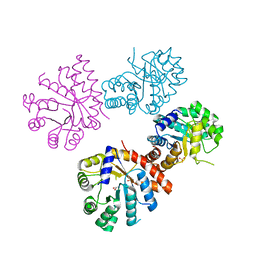 | |
5GUK
 
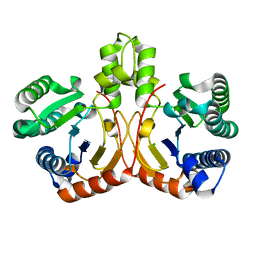 | | Crystal structure of apo form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | CHLORIDE ION, Cyclolavandulyl diphosphate synthase | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3B2Q
 
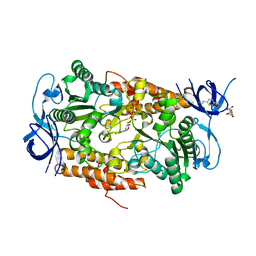 | | Intermediate position of ATP on its trail to the binding pocket inside the subunit B mutant R416W of the energy converter A1Ao ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, ... | | Authors: | Kumar, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Gruber, G. | | Deposit date: | 2007-10-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spectroscopic and crystallographic studies of the mutant R416W give insight into the nucleotide binding traits of subunit B of the A1Ao ATP synthase
Proteins, 75, 2009
|
|
2A58
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound riboflavin | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A57
 
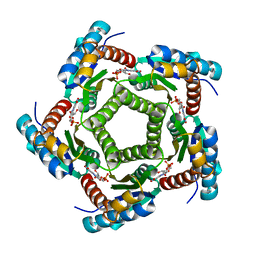 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
