7LR4
 
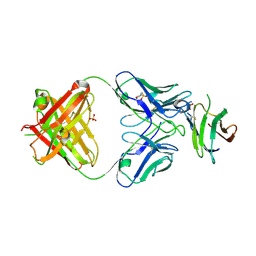 | | Complex of Fab 2/1.12 with domain 3 of P. berghei HAP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D3_2/1.12 Fab heavy chain, D3_2/1.12 Fab light chain, ... | | Authors: | Feng, J, Dong, X.C, Su, Y, Lu, X.F, Springer, T.A. | | Deposit date: | 2021-02-15 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of malaria transmission blockade by a monoclonal antibody to gamete fusogen HAP2.
Elife, 10, 2021
|
|
4ETJ
 
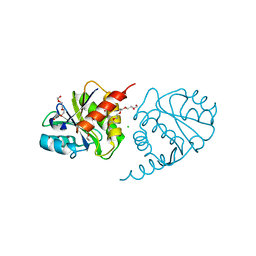 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6KAF
 
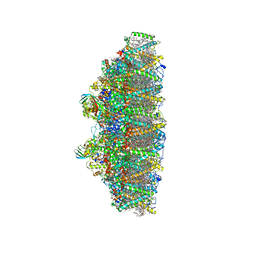 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KAD
 
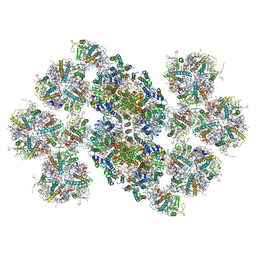 | | Cryo-EM structure of the C2S2M2L2-type PSII-LHCII supercomplex from Chlamydomonas reihardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Sheng, X, Watanabe, A, Li, A.J, Kim, E, Song, C, Murata, K, Song, D.F, Minagawa, J, Liu, Z.F. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into light harvesting for photosystem II in green algae.
Nat.Plants, 5, 2019
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
5MRD
 
 | | Human PDK1-PKCiota Kinase Chimera in Complex with Allosteric Compound PS267 Bound to the PIF-Pocket | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Arencibia, J.M, Froehner, W, Krupa, M, Pastor-Flores, D, Merker, P, Oellerich, T, Neimanis, S, Schmithals, C, Koeberle, V, Suess, E, Zeuzem, S, Stark, H, Piiper, A, Odadzic, D, Schulze, J.O, Biondi, R.M. | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | An Allosteric Inhibitor Scaffold Targeting the PIF-Pocket of Atypical Protein Kinase C Isoforms.
ACS Chem. Biol., 12, 2017
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
6JQH
 
 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
6BKA
 
 | | Crystal Structure of Nitronate Monooxygenase from Cyberlindnera saturnus | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Agniswamy, J, Fang, Y.-F, Weber, I.T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yeast nitronate monooxygenase from Cyberlindnera saturnus.
Proteins, 86, 2018
|
|
8H41
 
 | | Crystal structure of a decarboxylase from Trichosporon moniliiforme in complex with o-nitrophenol | | Descriptor: | MAGNESIUM ION, O-NITROPHENOL, Salicylate decarboxylase | | Authors: | Gao, J, Zhao, Y.P, Li, Q, Liu, W.D, Sheng, X. | | Deposit date: | 2022-10-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Combined Computational-Experimental Study on the Substrate Binding and Reaction Mechanism of Salicylic Acid Decarboxylase
Catalysts, 12, 2022
|
|
6KTS
 
 | | Structure of C34N126K/N36 | | Descriptor: | Envelope glycoprotein, Glycoprotein 41 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Characterization of the Secondary Mutation N126K Selected by Various HIV-1 Fusion Inhibitors.
Viruses, 12, 2020
|
|
6BG9
 
 | |
5WVC
 
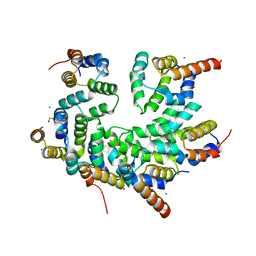 | | Structure of the CARD-CARD disk | | Descriptor: | Apoptotic protease-activating factor 1, Caspase, IODIDE ION | | Authors: | Lin, S.C, Lo, Y.C, Su, T.W. | | Deposit date: | 2016-12-24 | | Release date: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Structural Insights into DD-Fold Assembly and Caspase-9 Activation by the Apaf-1 Apoptosome.
Structure, 25, 2017
|
|
8UHD
 
 | |
8UIS
 
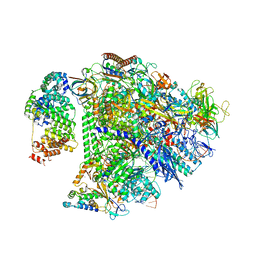 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
2HQD
 
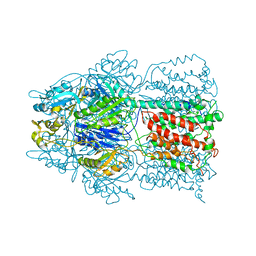 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
8THK
 
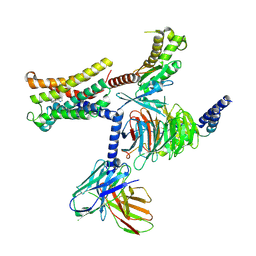 | |
8THL
 
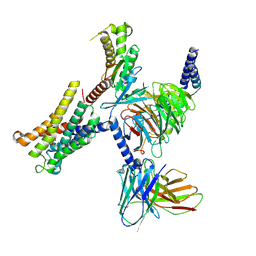 | |
8EMT
 
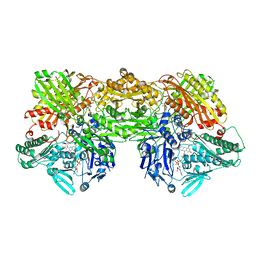 | | Cryo-EM analysis of the human aldehyde oxidase from liver | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
2HQF
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQC
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQG
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
4RCB
 
 | | Crystal structure of E Coli Hfq | | Descriptor: | RNA-binding protein Hfq | | Authors: | Su, J.Y. | | Deposit date: | 2014-09-15 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Limited proteolysis improves E.coli Hfq crystal structure resolution
Chin J Biochem Mol Biol, 9, 2015
|
|
4RCC
 
 | | Crystal structure of E Coli Hfq | | Descriptor: | RNA-binding protein Hfq | | Authors: | Su, J.Y. | | Deposit date: | 2014-09-15 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Limited proteolysis improves E.coli Hfq crystal structure resolution
Chin J Biochem Mol Biol, 9, 2015
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
