7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | 著者 | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | 登録日 | 2020-05-07 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7BW4
 
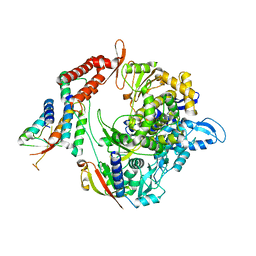 | | Structure of the RNA-dependent RNA polymerase from SARS-CoV-2 | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | 著者 | Peng, Q, Peng, R, Shi, Y. | | 登録日 | 2020-04-13 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Cell Rep, 31, 2020
|
|
4NYZ
 
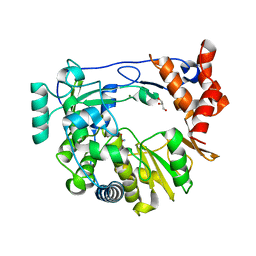 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | 分子名称: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | 著者 | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | 登録日 | 2013-12-11 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
4NZ0
 
 | |
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | 分子名称: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | 登録日 | 2020-09-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4Y2A
 
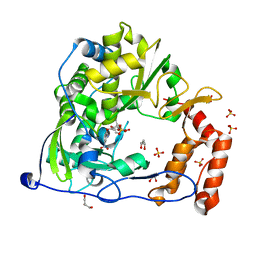 | | Crystal Structure of Coxsackie Virus B3 3D polymerase in complex with GPC-N114 inhibitor | | 分子名称: | 2,2'-[(4-chlorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | 著者 | Vives-Adrian, L, Ferrer-Orta, C, Verdaguer, N. | | 登録日 | 2015-02-09 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
4Y3C
 
 | |
4Y2C
 
 | |
1RA7
 
 | | Poliovirus Polymerase with GTP | | 分子名称: | ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | 著者 | Thompson, A.A, Peersen, O.B. | | 登録日 | 2003-10-31 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase.
Embo J., 23, 2004
|
|
1RAJ
 
 | |
1RA6
 
 | |
4KB7
 
 | | HCV NS5B GT1B N316Y with CMPD 32 | | 分子名称: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[{2-[(3R)-1-hydroxy-1,3-dihydro-2,1-benzoxaborol-3-yl]ethyl}(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, HCV Polymerase | | 著者 | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | 登録日 | 2013-04-23 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
4KE5
 
 | | HCV NS5B GT1B N316Y with GSK5852 | | 分子名称: | HCV Polymerase, [4-({[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}methyl)-2-fluorophenyl]boronic acid | | 著者 | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | 登録日 | 2013-04-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
4KBI
 
 | | HCV NS5B GT1B N316Y with CMPD 4 | | 分子名称: | 5-cyclopropyl-6-{[(7-fluoro-1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)methyl](methylsulfonyl)amino}-2-(4-fluorophenyl)-N-methyl-1-benzofuran-3-carboxamide, HCV Polymerase | | 著者 | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | 登録日 | 2013-04-23 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
1RDR
 
 | |
4RY6
 
 | |
4RY4
 
 | |
4RY7
 
 | |
4RY5
 
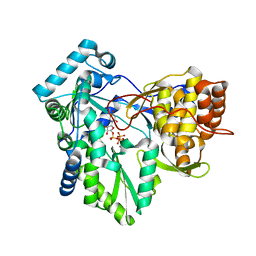 | | C-terminal mutant (W550N) of HCV/J4 RNA polymerase | | 分子名称: | HCV J4 RNA polymerase (NS5B), MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE | | 著者 | Jaeger, J, Cherry, A, Dennis, C. | | 登録日 | 2014-12-13 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
6L4R
 
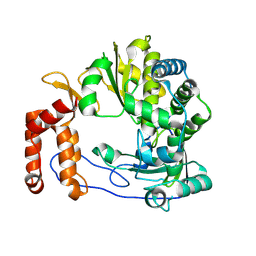 | | Crystal structure of Enterovirus D68 RdRp | | 分子名称: | RdRp | | 著者 | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | 登録日 | 2019-10-21 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
2J7N
 
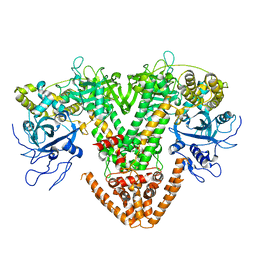 | | Structure of the RNAi polymerase from Neurospora crassa | | 分子名称: | GLYCEROL, MAGNESIUM ION, RNA-DEPENDENT RNA POLYMERASE | | 著者 | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | 登録日 | 2006-10-13 | | 公開日 | 2006-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
4KHM
 
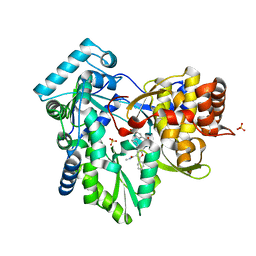 | | HCV NS5B GT1A with GSK5852 | | 分子名称: | HCV Polymerase, SULFATE ION, [4-({[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}methyl)-2-fluorophenyl]boronic acid | | 著者 | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | 登録日 | 2013-04-30 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
4KAI
 
 | | HCV NS5B GT1B N316 with GSK5852A | | 分子名称: | HCV Polymerase, [4-({[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}methyl)-2-fluorophenyl]boronic acid | | 著者 | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | 登録日 | 2013-04-22 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
7B3D
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | 分子名称: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | 著者 | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | 登録日 | 2020-11-30 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
2R92
 
 | | Elongation complex of RNA polymerase II with artificial RdRP scaffold | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Lehmann, E, Brueckner, F, Cramer, P. | | 登録日 | 2007-09-12 | | 公開日 | 2007-11-27 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Molecular basis of RNA-dependent RNA polymerase II activity.
Nature, 450, 2007
|
|
