6MEA
 
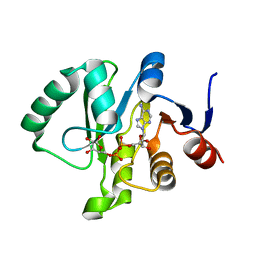 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEN
 
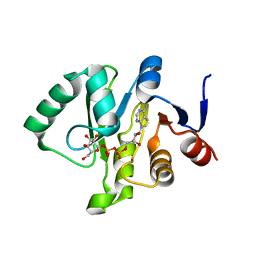 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6FX7
 
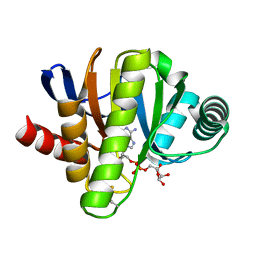 | | Crystal structure of in vitro evolved Af1521 | | Descriptor: | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, [Protein ADP-ribosylglutamate] hydrolase AF_1521 | | Authors: | Karlberg, T, Thorsell, A.G, Nowak, K, Hottiger, M.O, Schuler, H. | | Deposit date: | 2018-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of in vitro evolved Af1521
To Be Published
|
|
6VXS
 
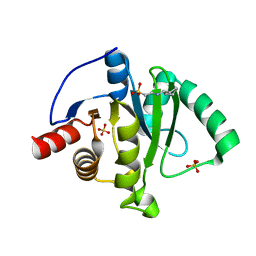 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Non-structural protein 3, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W02
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6QZU
 
 | | Getah virus macro domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0G
 
 | | Getah virus macro domain in complex with ADPr, pose 2 | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural polyprotein | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0P
 
 | | Getah virus macro domain in complex with ADPr in double open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0F
 
 | | Getah virus macro domain in complex with ADPr, pose 1 | | Descriptor: | Non-structural polyprotein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0T
 
 | | Getah virus macro domain in complex with ADPr in open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0R
 
 | | Getah virus macro domain in complex with ADPr covalently bond to Cys34 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural polyprotein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S})-2,3,4,5-tetrakis(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6WCF
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.065 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WEN
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | Descriptor: | CHLORIDE ION, Non-structural protein 3 | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WEY
 
 | |
6YWK
 
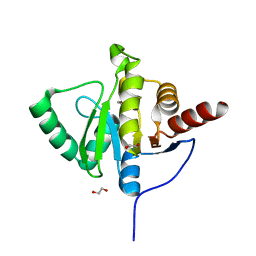 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YWM
 
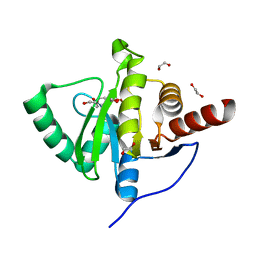 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6WOJ
 
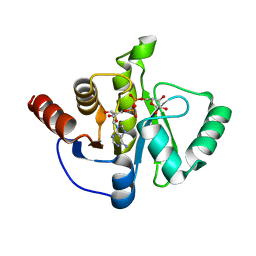 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
7JME
 
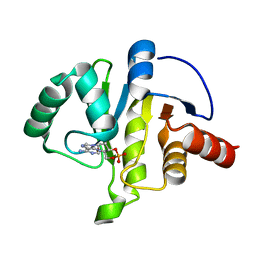 | |
6Y4Z
 
 | | The crystal structure of human MACROD2 in space group P43212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y4Y
 
 | | The crystal structure of human MACROD2 in space group P41212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y73
 
 | | The crystal structure of human MACROD2 in space group P43 | | Descriptor: | ADP-ribose glycohydrolase MACROD2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7D2C
 
 | | The Crystal Structure of human PARP14 from Biortus. | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Wang, F, Miao, Q, Lv, Z, Cheng, W, Lin, D, Xu, X, Tan, J. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Crystal Structure of human PARP14 from Biortus.
To Be Published
|
|
7KG3
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
