7TMG
 
 | |
7TMF
 
 | |
7T2Z
 
 | |
7T33
 
 | |
8CQV
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CJ0
 
 | | E. coli NfsB-T41Q/N71S/F124T/M127V mutant bound to nicotinate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, ... | | Authors: | White, S.A, Hyde, E.I, Day, M.A. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8C5P
 
 | | E. coli NfsB mutant N71S T41L with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.A, White, S.A, Hyde, E.I. | | Deposit date: | 2023-01-10 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8CCV
 
 | | E. coli NfsB mutant T41LN71S with nicotinate | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
4G8S
 
 | | Crystal Structure Of a Putative Nitroreductase from Geobacter sulfurreducens PCA (Target PSI-013445) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Nitroreductase family protein | | Authors: | Kumar, P.R, Bhosle, R, Hillerich, B, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nitroreductase from Geobacter sulfurreducens PCA
to be published
|
|
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQD
 
 | | Structure of Nitroreductase from E. cloacae Bound with 2e-Reduced Flavin Mononucleotide (FMN) | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQB
 
 | | Structure of Nitroreductase from E. cloacae complex with inhibitor benzoate | | Descriptor: | BENZOIC ACID, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1NEC
 
 | | NITROREDUCTASE FROM ENTEROBACTER CLOACAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (NITROREDUCTASE) | | Authors: | Hecht, H.J, Bryant, C, Erdmann, H, Pelletier, H, Sawaya, R. | | Deposit date: | 1999-03-30 | | Release date: | 2000-03-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Nitroreductase from Enterobacter Cloacae
To be Published
|
|
1NOX
 
 | | NADH OXIDASE FROM THERMUS THERMOPHILUS | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH OXIDASE | | Authors: | Hecht, H.J, Erdmann, H, Park, H.J, Sprinzl, M, Schmid, R.D. | | Deposit date: | 1996-11-20 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of NADH oxidase from Thermus thermophilus.
Nat.Struct.Biol., 2, 1995
|
|
1OON
 
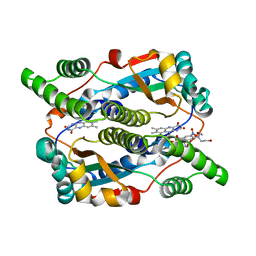 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN27217 | | Descriptor: | 2,4-DINITRO,5-[BIS(2-BROMOETHYL)AMINO]-N-(2',3'-DIOXOPROPYL)BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OO6
 
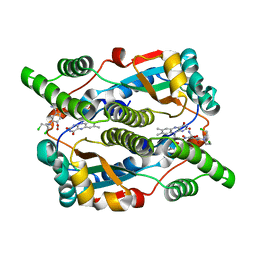 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN23862 | | Descriptor: | 5-[BIS-2(CHLORO-ETHYL)-AMINO]-2,4-DINTRO-BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-03 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OO5
 
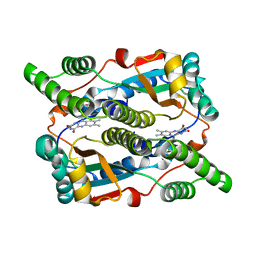 | | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of the Enzyme Active Form and Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-03 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OOQ
 
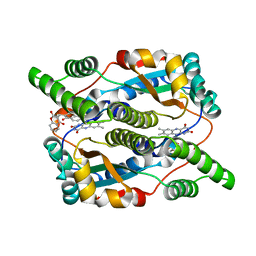 | | Nitroreductase from e-coli in complex with the inhibitor dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
3E10
 
 | |
3EO7
 
 | |
3E39
 
 | |
3EK3
 
 | |
3EO8
 
 | |
3EOF
 
 | |
3G14
 
 | |
