8SKN
 
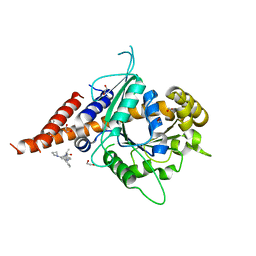 | | Crystal structure of compound 3-bound human Dynamin-1-like protein GTPase-BSE fusion | | Descriptor: | 1,2-ETHANEDIOL, Dynamin-1-like protein GTPase-BSE fusion, N-[4-(azetidin-1-yl)-2-(4-methylphenyl)quinolin-6-yl]-2-methylpropanamide | | Authors: | Ma, B. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Potent Allosteric DRP1 Inhibitors by Disrupting Protein-Protein Interaction with MiD49.
Acs Med.Chem.Lett., 14, 2023
|
|
4WHJ
 
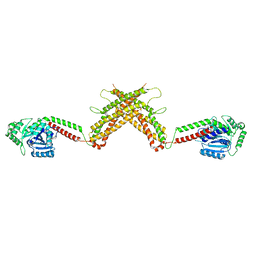 | |
8T1H
 
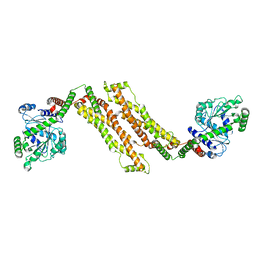 | |
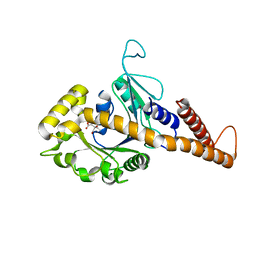
5UOT
 
 | |
5MTV
 
 | | Active structure of EHD4 complexed with ATP-gamma-S | | Descriptor: | EH domain-containing protein 4, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4H1U
 
 | | Nucleotide-free human dynamin-1-like protein GTPase-GED fusion | | Descriptor: | CITRATE ANION, Dynamin-1-like protein | | Authors: | Wenger, J, Klinglmayr, E, Puehringer, S, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
5MVF
 
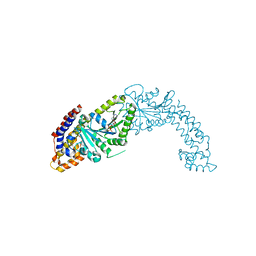 | | Active structure of EHD4 complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EH domain-containing protein 4, MAGNESIUM ION | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.268 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4H1V
 
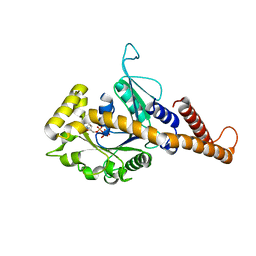 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | Descriptor: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
2J69
 
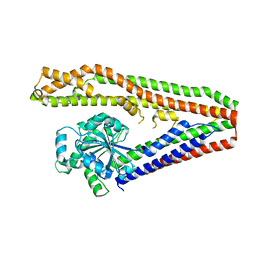 | | Bacterial dynamin-like protein BDLP | | Descriptor: | BACTERIAL DYNAMIN-LIKE PROTEIN | | Authors: | Low, H.H, Lowe, J. | | Deposit date: | 2006-09-26 | | Release date: | 2006-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Bacterial Dynamin-Like Protein
Nature, 444, 2006
|
|
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
2J68
 
 | |
2W6D
 
 | | BACTERIAL DYNAMIN-LIKE PROTEIN LIPID TUBE BOUND | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DYNAMIN FAMILY PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Low, H.H, Sachse, C, Amos, L.A, Lowe, J. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of a Bacterial Dynamin-Like Protein Lipid Tube Provides a Mechanism for Assembly and Membrane Curving.
Cell(Cambridge,Mass.), 139, 2009
|
|
2X2E
 
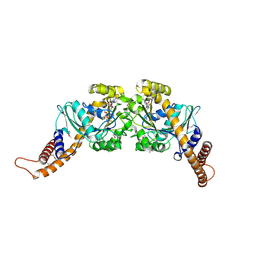 | | Dynamin GTPase dimer, long axis form | | Descriptor: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | Deposit date: | 2010-01-12 | | Release date: | 2010-04-28 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
2X2F
 
 | | Dynamin 1 GTPase dimer, short axis form | | Descriptor: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
4P4T
 
 | | GDP-bound stalkless-MxA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Interferon-induced GTP-binding protein Mx1, SULFATE ION | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
4P4S
 
 | | GMPPCP-bound stalkless-MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
4P4U
 
 | | Nucleotide-free stalkless-MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1, SULFATE ION | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
3ZYC
 
 | | DYNAMIN 1 GTPASE GED FUSION DIMER COMPLEXED WITH GMPPCP | | Descriptor: | DYNAMIN-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
4BEJ
 
 | | Nucleotide-free Dynamin 1-like protein (DNM1L, DRP1, DLP1) | | Descriptor: | DYNAMIN 1-LIKE PROTEIN | | Authors: | Froehlich, C, Schwefel, D, Faelber, K, Daumke, O. | | Deposit date: | 2013-03-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.483 Å) | | Cite: | Structural Insights Into Oligomerization and Mitochondrial Remodelling of Dynamin 1-Like Protein.
Embo J., 32, 2013
|
|
4CID
 
 | |
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZT
 
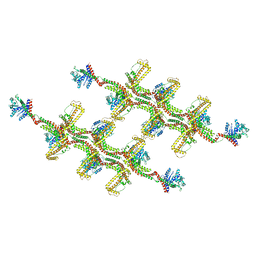 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZV
 
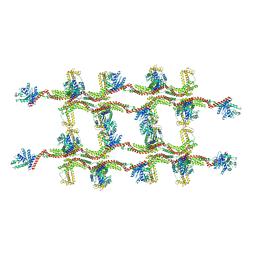 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (20.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZW
 
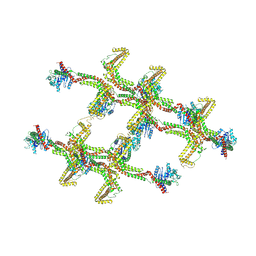 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6S9A
 
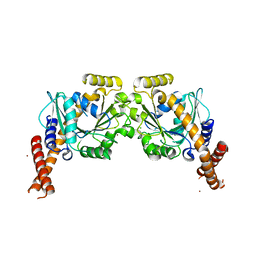 | | Artificial GTPase-BSE dimer of human Dynamin1 | | Descriptor: | CHLORIDE ION, Dynamin-1,Dynamin-1, ZINC ION | | Authors: | Ganichkin, O.M, Vancraenenbroeck, R, Rosenblum, G, Hofmann, H, Daumke, O, Noel, J.K. | | Deposit date: | 2019-07-11 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Quantification and demonstration of the collective constriction-by-ratchet mechanism in the dynamin molecular motor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
