5O5C
 
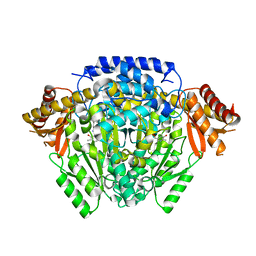 | | The crystal structure of DfoJ, the desferrioxamine biosynthetic pathway lysine decarboxylase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
3MAU
 
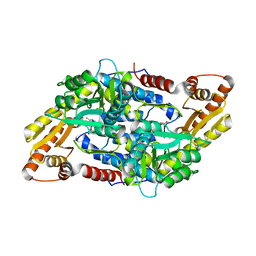 | | Crystal structure of StSPL in complex with phosphoethanolamine | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, sphingosine-1-phosphate lyase, ... | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-24 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3VP6
 
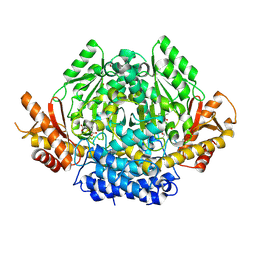 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | Descriptor: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | Authors: | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K1R
 
 | |
4E1O
 
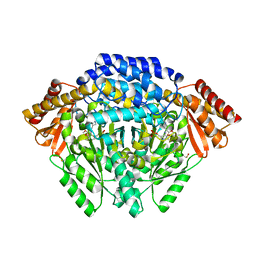 | | Human histidine decarboxylase complex with Histidine methyl ester (HME) | | Descriptor: | HISTIDINE-METHYL-ESTER, Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Komori, H, Nitta, Y, Ueno, H, Higuchi, Y. | | Deposit date: | 2012-03-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
J.Biol.Chem., 287, 2012
|
|
3K40
 
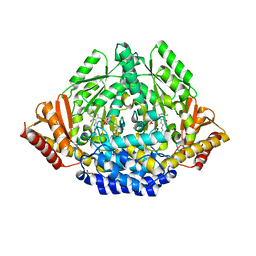 | | Crystal structure of Drosophila 3,4-dihydroxyphenylalanine decarboxylase | | Descriptor: | Aromatic-L-amino-acid decarboxylase, GLYCEROL | | Authors: | Han, Q, Ding, H, Robinson, H, Christensen, B.M, Li, J. | | Deposit date: | 2009-10-05 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase
Plos One, 5, 2010
|
|
2JIS
 
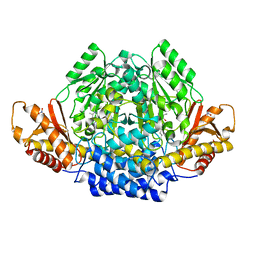 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | Descriptor: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-28 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
1XEY
 
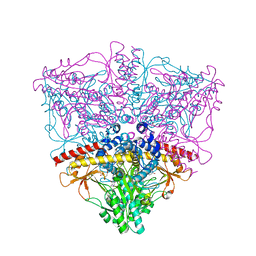 | | Crystal structure of the complex of Escherichia coli GADA with glutarate at 2.05 A resolution | | Descriptor: | ACETATE ION, GLUTARIC ACID, Glutamate decarboxylase alpha, ... | | Authors: | Dutyshev, D.I, Darii, E.L, Fomenkova, N.P, Pechik, I.V, Polyakov, K.M, Nikonov, S.V, Andreeva, N.S, Sukhareva, B.S. | | Deposit date: | 2004-09-13 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli glutamate decarboxylase (GADalpha) in complex with glutarate at 2.05 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1JS6
 
 | | Crystal Structure of DOPA decarboxylase | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Dominici, P, Borri-Voltattorni, C, Jansonius, J.N, Malashkevich, V.N. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
Nat.Struct.Biol., 8, 2001
|
|
8AYF
 
 | | Crystal structure of human Sphingosine-1-phosphate lyase 1 | | Descriptor: | ACETATE ION, GLYCEROL, Sphingosine-1-phosphate lyase 1 | | Authors: | Giardina, G, Catalano, F, Pampalone, G, Cellini, B. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Sci Rep, 13, 2023
|
|
6ZEK
 
 | | Crystal structure of mouse CSAD | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Cysteine sulfinic acid decarboxylase, ... | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J.Struct.Biol., 213, 2021
|
|
7A0A
 
 | | Crystal structure of mouse CSAD in apo form | | Descriptor: | Cysteine sulfinic acid decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of cysteine sulphinic acid decarboxylase reveals structural determinants for substrate specificity of pyridoxal phosphate-dependent decarboxylases
To be published
|
|
8CMX
 
 | |
4OBV
 
 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | Descriptor: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Fraser, J.S, Van Benschoten, A.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
1JS3
 
 | | Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa | | Descriptor: | CARBIDOPA, DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Burkhard, P, Dominici, P, Borri-Voltattorni, C, Jansonius, J.N, Malashkevich, V.N. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
Nat.Struct.Biol., 8, 2001
|
|
1PMM
 
 | | Crystal structure of Escherichia coli GadB (low pH) | | Descriptor: | ACETIC ACID, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PMO
 
 | | Crystal structure of Escherichia coli GadB (neutral pH) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
4OBU
 
 | |
7ERV
 
 | | Crystal structure of L-histidine decarboxylase (C57S/C101V/C282V mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase, IMIDAZOLE | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
7ERU
 
 | | Crystal structure of L-histidine decarboxylase (C57S mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
2DGL
 
 | | Crystal structure of Escherichia coli GadB in complex with bromide | | Descriptor: | ACETIC ACID, BROMIDE ION, Glutamate decarboxylase beta, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
3RBL
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | CHLORIDE ION, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RCH
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the open conformation with LLP and PLP bound to Chain-A and Chain-B respectively | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-31 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2DGM
 
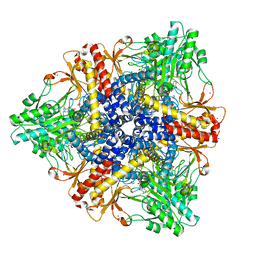 | | Crystal structure of Escherichia coli GadB in complex with iodide | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
