3P4X
 
 | | Helicase domain of reverse gyrase from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-10-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Conformational Flexibility of the Helicase-like Domain from Thermotoga maritima Reverse Gyrase Is Restricted by the Topoisomerase Domain.
Biochemistry, 50, 2011
|
|
3PEW
 
 | | S. cerevisiae Dbp5 L327V bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEY
 
 | | S. cerevisiae Dbp5 bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3RRN
 
 | | S. cerevisiae dbp5 l327v bound to gle1 h337r and ip6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3RRM
 
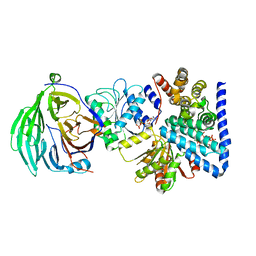 | | S. cerevisiae dbp5 l327v bound to nup159, gle1 h337r, ip6 and adp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
2YKG
 
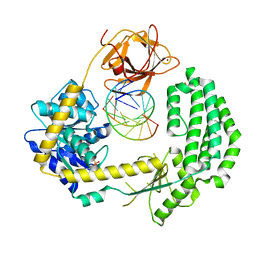 | | Structural insights into RNA recognition by RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
3SQX
 
 | |
3SQW
 
 | |
3TBK
 
 | | Mouse RIG-I ATPase Domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RIG-I Helicase Domain | | Authors: | Civril, F, Bennett, M.D, Hopfner, K.-P. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling.
Embo Rep., 12, 2011
|
|
4A2Q
 
 | | Structure of duck RIG-I tandem CARDs and helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A2P
 
 | | Structure of duck RIG-I helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A2W
 
 | | Structure of full-length duck RIG-I | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A36
 
 | | Structure of duck RIG-I helicase domain bound to 19-mer dsRNA and ATP transition state analogue | | Descriptor: | 5'-R(*GP*CP*AP*UP*GP*CP*GP*AP*CP*CP*UP*CP*UP*GP *UP*UP*UP*GP*A)-3', 5'-R(*UP*CP*AP*AP*AP*CP*AP*GP*AP*GP*GP*UP*CP*GP *CP*AP*UP*GP*C)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A4D
 
 | | Crystal structure of the N-terminal domain of the Human DEAD-BOX RNA helicase DDX5 (P68) | | Descriptor: | PROBABLE ATP-DEPENDENT RNA HELICASE DDX5 | | Authors: | Dutta, S, Choi, Y.W, Kotaka, M, Fielding, B.C, Tan, Y.J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Variable N-Terminal Region of Ddx5 Contains Structural Elements and Auto-Inhibits its Interaction with Ns5B of Hepatitis C Virus.
Biochem.J., 446, 2012
|
|
4A4Z
 
 | | CRYSTAL STRUCTURE OF THE S. CEREVISIAE DEXH HELICASE SKI2 BOUND TO AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, ANTIVIRAL HELICASE SKI2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Halbach, F, Rode, M, Conti, E. | | Deposit date: | 2011-10-20 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of S. Cerevisiae Ski2, a Dexh Helicase Associated with the Cytoplasmic Functions of the Exosome.
RNA, 18, 2012
|
|
4DDX
 
 | |
4DDU
 
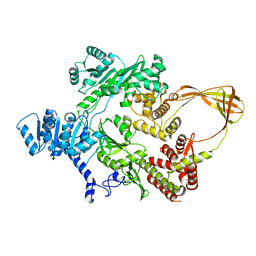 | | Thermotoga maritima reverse gyrase, C2 FORM 1 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDT
 
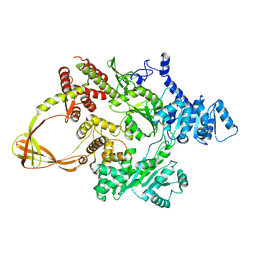 | | Thermotoga maritima reverse gyrase, C2 FORM 2 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDW
 
 | |
4DDV
 
 | |
4F92
 
 | | Brr2 Helicase Region S1087L | | Descriptor: | SULFANILAMIDE, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F91
 
 | | Brr2 Helicase Region | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F93
 
 | | Brr2 Helicase Region S1087L, Mg-ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AY2
 
 | | Capturing 5' tri-phosphorylated RNA duplex by RIG-I | | Descriptor: | 5'-R-PPP(GP*GP*CP*GP*CP*GP*GP*CP*UP*UP*CP*GP*GP*CP *CP*GP*CP*GP*CP*C)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-06-17 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing the Determinants of Viral RNA Recognition by Innate Immune Sensor Rig-I.
Structure, 20, 2012
|
|
3ZD7
 
 | | Snapshot 3 of RIG-I scanning on RNA duplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-11-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defining the Functional Determinants for RNA Surveillance by Rig-I.
Embo Rep., 14, 2013
|
|
