2MBE
 
 | |
2MJ5
 
 | | Structure of the UBA Domain of Human NBR1 in Complex with Ubiquitin | | Descriptor: | Next to BRCA1 gene 1 protein, Polyubiquitin-C | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-12-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
2LWP
 
 | |
2LXA
 
 | |
2MOR
 
 | |
2MRE
 
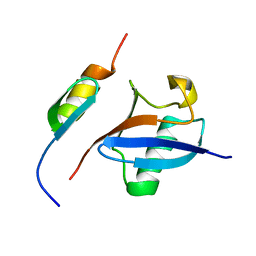 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2N2K
 
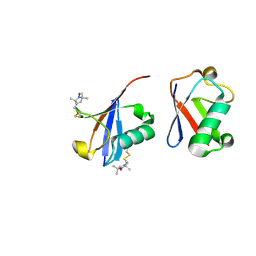 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-09-23 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
2N7K
 
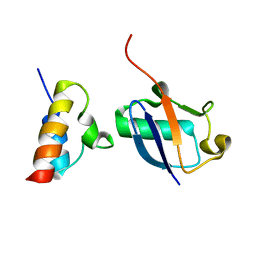 | | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8 | | Descriptor: | NEDD8, Protein KHNYN | | Authors: | Santonico, E, Nepravishta, R, Mattioni, A, Valentini, E, Mandaliti, W, Procopio, R, Iannuccelli, M, Castagnoli, L, Polo, S, Paci, M, Cesareni, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8
To be Published
|
|
2MSG
 
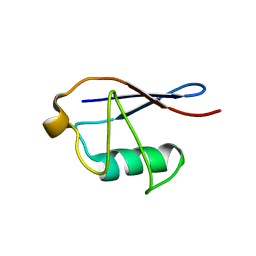 | | Solid-state NMR structure of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Lakomek, N, Habenstein, B, Loquet, A, Shi, C, Giller, K, Wolff, S, Becker, S, Fasshuber, H, Lange, A. | | Deposit date: | 2014-08-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural heterogeneity in microcrystalline ubiquitin studied by solid-state NMR.
Protein Sci., 24, 2015
|
|
2MWS
 
 | |
2NBD
 
 | |
2NBE
 
 | |
2N13
 
 | |
2N7D
 
 | |
2MQJ
 
 | |
2NBU
 
 | |
2N4F
 
 | | EC-NMR Structure of Arabidopsis thaliana At2g32350 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein AR3433A | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2OJR
 
 | |
2O6V
 
 | |
2NR2
 
 | | The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native states ensembles of proteins | | Descriptor: | Ubiquitin | | Authors: | Richter, B, Gsponer, J, Varnai, P, Salvatella, X, Vendruscolo, M. | | Deposit date: | 2006-11-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native state ensembles of proteins
J.Biomol.Nmr, 37, 2007
|
|
1CMX
 
 | |
1EUV
 
 | |
2QHO
 
 | |
2RR9
 
 | | The solution structure of the K63-Ub2:tUIMs complex | | Descriptor: | Putative uncharacterized protein UIMC1, ubiquitin | | Authors: | Sekiyama, N, Jee, J, Isogai, S, Akagi, K, Huang, T, Ariyoshi, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the K63-Ub2:tUIMs complex
To be Published
|
|
2WDT
 
 | | Crystal structure of Plasmodium falciparum UCHL3 in complex with the suicide inhibitor UbVME | | Descriptor: | CHLORIDE ION, UBIQUITIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE L3 | | Authors: | Weihofen, W.A, Artavanis-Tsakonas, K, Gaudet, R, Ploegh, H.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and Structural Studies of the Plasmodium Falciparum Ubiquitin and Nedd8 Hydrolase Uchl3.
J.Biol.Chem., 285, 2010
|
|
