4TV7
 
 | |
4CVQ
 
 | | CRYSTAL STRUCTURE OF AN AMINOTRANSFERASE FROM ESCHERICHIA COLI AT 2. 11 ANGSTROEM RESOLUTION | | Descriptor: | ACETATE ION, GLUTAMATE-PYRUVATE AMINOTRANSFERASE ALAA, GLYCEROL, ... | | Authors: | Penya-Soler, E, Fernandez, F.J, Lopez-Estepa, M, Garces, F, Richardson, A.J, Rudd, K.E, Coll, M, Vega, M.C. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural analysis and mutant growth properties reveal distinctive enzymatic and cellular roles for the three major L-alanine transaminases of Escherichia coli.
PLoS ONE, 9, 2014
|
|
4N0B
 
 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MY5
 
 | | Crystal structure of the aromatic amino acid aminotransferase from Streptococcus mutants | | Descriptor: | Putative amino acid aminotransferase | | Authors: | Cong, X, Li, X, Ge, J, Feng, Y, Feng, X, Li, S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of the aromatic-amino-acid aminotransferase from Streptococcus mutans.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4MGR
 
 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M2M
 
 | | Crystal structure of PLP-dependent cyclase OrfR in complex with PLP-L-Arg | | Descriptor: | ARGININE, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-08-05 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4M2K
 
 | | Crystal structure of PLP-dependent cyclase OrfR in complex with PLP | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-08-05 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4M2J
 
 | | Crystal structure of PLP-dependent cyclase OrfR in complex with Au | | Descriptor: | Aminotransferase, GOLD ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-08-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4BMK
 
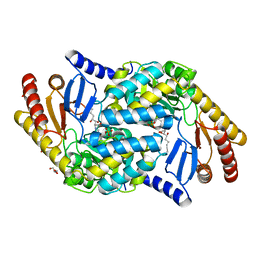 | | Serine Palmitoyltransferase K265A from S. paucimobilis with bound PLP- Myriocin Aldimine | | Descriptor: | Decarboxylated Myriocin, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wadsworth, J.M, Clarke, D.J, McMahon, S.A, Beattie, A.E, Lowther, J, Dunn, T.M, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Chemical Basis of Serine Palmitoyltransferase Inhibition by Myriocin.
J.Am.Chem.Soc., 135, 2013
|
|
4JE5
 
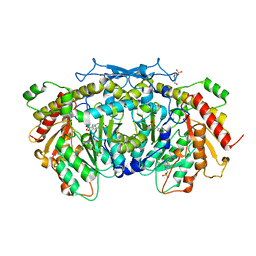 | | Crystal structure of the aromatic aminotransferase Aro8, a putative alpha-aminoadipate aminotransferase in Saccharomyces cerevisiae | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic/aminoadipate aminotransferase 1, ... | | Authors: | Bulfer, S.L, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2013-02-26 | | Release date: | 2013-09-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase.
Protein Sci., 22, 2013
|
|
4J8L
 
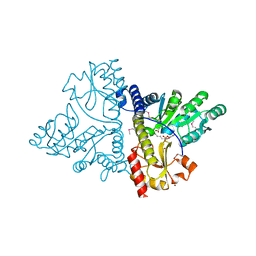 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT protein YhfS from Escherichia coli | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, NICKEL (II) ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of the pyridoxal-5'-phosphate dependent protein yhfs from escherichia coli
To be Published
|
|
4IX8
 
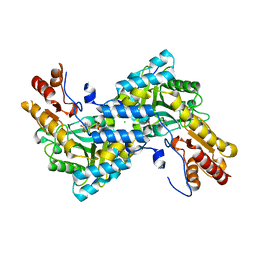 | |
4IW7
 
 | | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis. | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Newcomb, W, Niedzialkowska, E, Porebski, P.J, Grimshaw, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis.
To be Published
|
|
4H51
 
 | |
4GEB
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(7-hydroxy-6-oxo-2-phenyl-6,7-dihydro-2H-pyrazolo[3,4-b]pyridin-5-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial, ... | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PF-04859989 as a template for structure-based drug design: identification of new pyrazole series of irreversible KAT II inhibitors with improved lipophilic efficiency.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4GE7
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-2-oxo-6-phenoxy-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Irreversible Human KAT II Inhibitors: Discovery of New Potency-Enhancing Interactions.
ACS Med Chem Lett, 4, 2013
|
|
4GE4
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-7-methoxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Irreversible Human KAT II Inhibitors: Discovery of New Potency-Enhancing Interactions.
ACS Med Chem Lett, 4, 2013
|
|
4GDY
 
 | | Kynurenine Aminotransferase II inhibitors | | Descriptor: | (5-hydroxy-6-methyl-4-{[(1-oxo-7-phenoxy-1,2-dihydro[1,2,4]triazolo[4,3-a]quinolin-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of Hydroxamate Bioisosteres as KATII Inhibitors with Improved Oral Bioavailability and Pharmacokinetics
MEDCHEMCOMM, 2012
|
|
4GE9
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (4-{[(6-benzyl-1-hydroxy-7-methoxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Design of Irreversible Human KAT II Inhibitors: Discovery of New Potency-Enhancing Interactions.
ACS Med Chem Lett, 4, 2013
|
|
4FL0
 
 | | Crystal structure of ALD1 from Arabidopsis thaliana | | Descriptor: | Aminotransferase ALD1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sobolev, V, Edelman, M, Dym, O, Unger, T, Albeck, S, Kirma, M, Galili, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2012-06-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ALD1, a plant-specific homologue of the universal diaminopimelate aminotransferase enzyme of lysine biosynthesis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4F5J
 
 | |
4F5F
 
 | |
4F5M
 
 | |
4F5I
 
 | |
4F5H
 
 | |
