5T2E
 
 | |
2PWR
 
 | | HIV-1 protease in complex with a carbamoyl decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-{(3S,4S)-PYRROLIDINE-3,4-DIYLBIS[(BENZYLIMINO)SULFONYL]}DIBENZAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boettcher, B, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
5YOJ
 
 | | Structure of A17 HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, A17 HIV-1 protease, GLYCEROL | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
5AH6
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AHC
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AHB
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
4DQB
 
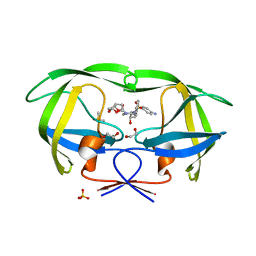 | | Crystal Structure of wild-type HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
7MYY
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
1XL2
 
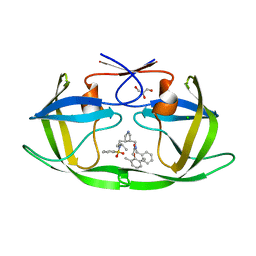 | | HIV-1 Protease in complex with pyrrolidinmethanamine | | Descriptor: | CHLORIDE ION, GLYCEROL, N-BENZYL-2-(2,6-DIMETHYLPHENOXY)-N-[((3R,4S)-4-{[ISOBUTYL(PHENYLSULFONYL)AMINO]METHYL}PYRROLIDIN-3-YL)METHYL]ACETAMIDE, ... | | Authors: | Boettcher, J, Specker, E, Heine, A, Klebe, G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Old Target Revisited: Two New Privileged Skeletons and an Unexpected Binding Mode For HIV-Protease Inhibitors
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
7MAS
 
 | | Drug Resistant HIV-1 Protease (L10F, M46I, I50V, F53L, L63P, G73S) in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug Resistant HIV-1 Protease (L10F, M46I, I50V, F53L, L63P, G73S) in Complex with DRV
To Be Published
|
|
4Q1Y
 
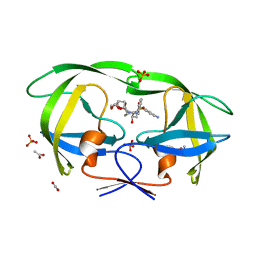 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
4QJ6
 
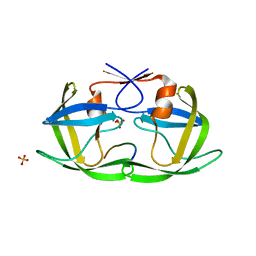 | |
2FGV
 
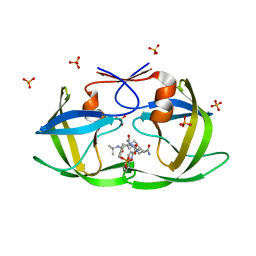 | | X-ray crystal structure of HIV-1 Protease T80N variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | Descriptor: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | Authors: | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | Deposit date: | 2005-12-22 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
4A6C
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | METHYL ((S)-1-(2-(3-((3S,4S)-3-BENZYL-4-HYDROXY-1-((1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL)-2-OXOPYRROLIDIN-3-YL)PROPYL)-2-(4-(PYRIDIN-4-YL)BENZYL)HYDRAZINYL)-3,3-DIMETHYL-1-OXOBUTAN-2-YL)CARBAMATE, POL PROTEIN | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Unge, J, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B.B, Unge, J, Larhed, M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
3S53
 
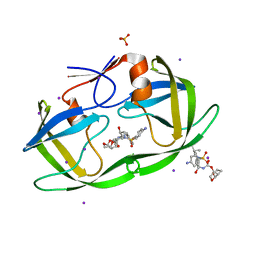 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P212121 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, IODIDE ION, PHOSPHATE ION, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
6B3G
 
 | |
3SM1
 
 | |
3SAC
 
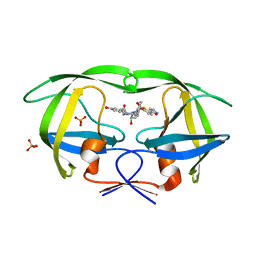 | | Crystal structure of wild-type HIV-1 protease in complex with AF80 | | Descriptor: | 3-hydroxy-N-{(2S,3R)-3-hydroxy-4-[(2-methylpropyl){[5-(1,2-oxazol-5-yl)thiophen-2-yl]sulfonyl}amino]-1-phenylbutan-2-yl}benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
5IVR
 
 | |
3SA7
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AF55 | | Descriptor: | N~2~-acetyl-N-[(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-L-leucinamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SA8
 
 | | Crystal structure of wild-type HIV-1 protease in complex with KB83 | | Descriptor: | 3-hydroxy-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]benzamide, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3B80
 
 | | HIV-1 protease mutant I54V complexed with gem-diol-amine intermediate NLLTQI | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Chumanevich, A.A, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in the Act: The 1.5 A Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate.
Biochemistry, 46, 2007
|
|
3SAB
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AF78 | | Descriptor: | 3-hydroxy-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](3-phenylpropyl)amino}-1-phenylbutan-2-yl]benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
6OTG
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Pawar, S, Weber, I.T. | | Deposit date: | 2019-05-03 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of antiviral inhibitor with HIV-1 protease bearing drug resistant substitutions of V32I, I47V and V82I.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
