8RAS
 
 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8V4J
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8R8D
 
 | |
8X8T
 
 | |
8X83
 
 | |
8V3B
 
 | |
8R7M
 
 | | CTX-M14 in complex with boric acid and 1,2-diol boric ester | | Descriptor: | BORIC ACID, Beta-lactamase, GLYCEROL, ... | | Authors: | Werner, N, Prester, A, Hinrichs, W, Perbandt, M, Betzel, C. | | Deposit date: | 2023-11-26 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the Beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification
To Be Published
|
|
8V2T
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148591 | | Descriptor: | 1,5,6-trideoxy-6,6-difluoro-1-(N-hydroxyformamido)-6-phosphono-D-ribo-hexitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
8V2D
 
 | | Computational Designed Nanocage O43_129 | | Descriptor: | O43_129 component A, O43_129 component B | | Authors: | Weidle, C, Kibler, R.D. | | Deposit date: | 2023-11-22 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | Blueprinting extendable nanomaterials with standardized protein blocks.
Nature, 627, 2024
|
|
8R6F
 
 | | CryoEM structure of wheat 40S ribosomal subunit, body domain | | Descriptor: | 30S ribosomal protein S11, chloroplastic, 30S ribosomal protein S4, ... | | Authors: | Kravchenko, O.V, Baymukhametov, T.N, Afonina, Z.A, Vasilenko, K.S. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | High-Resolution Structure and Internal Mobility of a Plant 40S Ribosomal Subunit.
Int J Mol Sci, 24, 2023
|
|
8R6S
 
 | | Plastid-encoded RNA polymerase (Integrated model) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-22 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8V1F
 
 | | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Halabelian, L, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol
To Be Published
|
|
8X5L
 
 | | The Crystal Structure of PRKACA from Biortus. | | Descriptor: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, SODIUM ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of PRKACA from Biortus.
To Be Published
|
|
8R5O
 
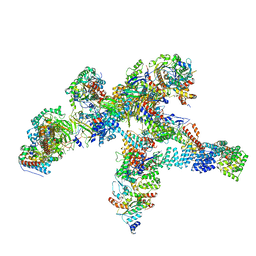 | | Plastid-encoded RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8R57
 
 | | CryoEM structure of wheat 40S ribosomal subunit, head domain | | Descriptor: | 40S ribosomal protein S12, 40S ribosomal protein S15, 40S ribosomal protein S16, ... | | Authors: | Kravchenko, O.V, Baymukhametov, T.N, Afonina, Z.A, Vasilenko, K.S. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | High-Resolution Structure and Internal Mobility of a Plant 40S Ribosomal Subunit.
Int J Mol Sci, 24, 2023
|
|
8R55
 
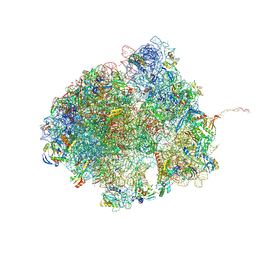 | | Bacillus subtilis MutS2-collided disome complex (collided 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S RNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
8X4F
 
 | |
8UZN
 
 | |
8UZM
 
 | |
8UZO
 
 | |
8UZK
 
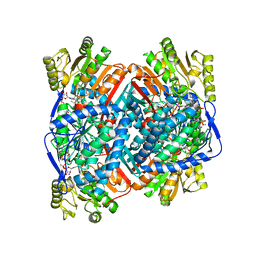 | |
8UZJ
 
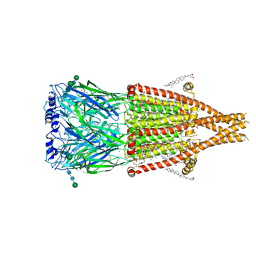 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and ivermectin | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
7GS1
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-24 | | Descriptor: | 2-cyano-~{N}-cyclohexyl-ethanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRI
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-5 | | Descriptor: | (1S)-1-(1H-pyrazol-5-yl)ethan-1-ol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
