7ZR5
 
 | |
7Z38
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7XNC
 
 | | MEK1 bound to DS94070624 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Kishikawa, S, Takano, K, Ubukata, O, Hanzawa, H. | | Deposit date: | 2022-04-28 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel ATP-Competitive MEK Inhibitor DS03090629 that Overcomes Resistance Conferred by BRAF Overexpression in BRAF-Mutated Melanoma.
Mol.Cancer Ther., 22, 2023
|
|
7XLP
 
 | | MEK1 bound to DS03090629 | | Descriptor: | (1~{R},3~{S})-3-[[6-[2-chloranyl-4-(4-methylpyrimidin-2-yl)oxy-phenyl]-3-methyl-1~{H}-indazol-4-yl]oxy]cyclohexan-1-amine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kishikawa, S, Takano, K, Ubukata, O, Hanzawa, H. | | Deposit date: | 2022-04-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel ATP-Competitive MEK Inhibitor DS03090629 that Overcomes Resistance Conferred by BRAF Overexpression in BRAF-Mutated Melanoma.
Mol.Cancer Ther., 22, 2023
|
|
7XC1
 
 | | Crystal structure of ERK2 with an allosteric inhibitor 3 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for ERK2 allosteric inhibitors.
To Be Published
|
|
7X4U
 
 | | Crystal structure of ERK2 with an allosteric inhibitor 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for producing allosteric ERK2 inhibitors
To Be Published
|
|
7W5O
 
 | | Crystal structure of ERK2 with an allosteric inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a novel target site for ATP-independent ERK2 inhibitors.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W4P
 
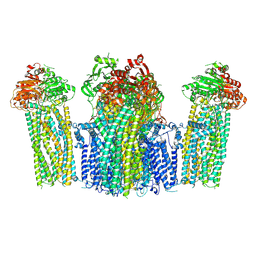 | | The structure of KATP H175K mutant in closed state | | Descriptor: | 6-chloranyl-~{N}-(1-methylcyclopropyl)-1,1-bis(oxidanylidene)-4~{H}-thieno[3,2-e][1,2,4]thiadiazin-3-amine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the mechanism of pancreatic K ATP channel regulation by nucleotides.
Nat Commun, 13, 2022
|
|
7W4O
 
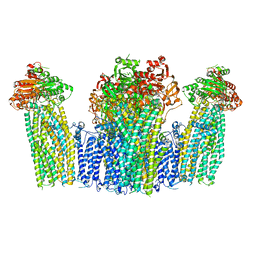 | | The structure of KATP H175K mutant in pre-open state | | Descriptor: | 6-chloranyl-~{N}-(1-methylcyclopropyl)-1,1-bis(oxidanylidene)-4~{H}-thieno[3,2-e][1,2,4]thiadiazin-3-amine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into the mechanism of pancreatic K ATP channel regulation by nucleotides.
Nat Commun, 13, 2022
|
|
7URT
 
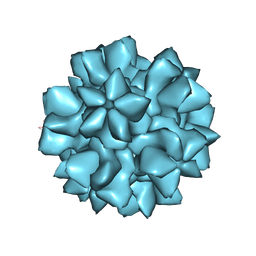 | |
7URN
 
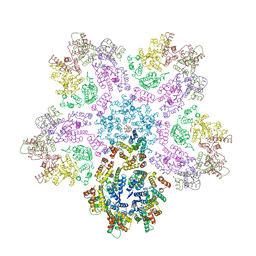 | |
7UP8
 
 | |
7UP7
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound with literature RSK2 inhibitor indazole cyanoacrylamide compound 26 (soak) | | Descriptor: | (2S)-2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]propanamide, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP6
 
 | | Crystal structure of C-terminal domain of MSK1 in complex with in covalently bound literature RSK2 inhibitor pyrrolopyrimidine cyanoacrylamide compound 25 (co-crystal) | | Descriptor: | (E)-3-(3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl)-2-cyanoacrylamide bound form, OXAMIC ACID, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP5
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 23 (co-crystal) | | Descriptor: | (2M)-6-chloro-2-(5H-pyrrolo[3,2-d]pyrimidin-5-yl)pyridine-3-carbonitrile, IODIDE ION, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Edwards, T.E, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP4
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 20 (co-crystal) | | Descriptor: | (5M)-5-(2,5-dichloropyrimidin-4-yl)-5H-pyrrolo[3,2-d]pyrimidine, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UMB
 
 | | NanoBRET tracer Tram-bo bound to a KSR2-MEK1 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Marsiglia, W.M, Khan, K.M, Dar, A.C. | | Deposit date: | 2022-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | Live-cell target engagement of allosteric MEKi on MEK-RAF/KSR-14-3-3 complexes.
Nat.Chem.Biol., 20, 2024
|
|
7UGB
 
 | |
7T13
 
 | |
7T12
 
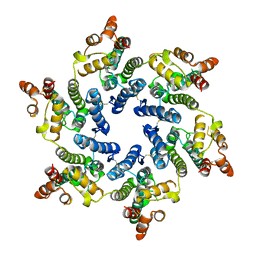 | | Hexameric HIV-1 (O-group) CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
7S61
 
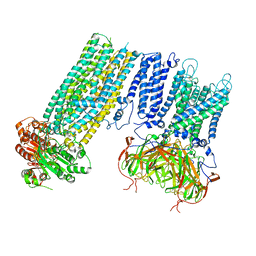 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S60
 
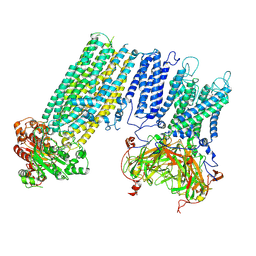 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Z
 
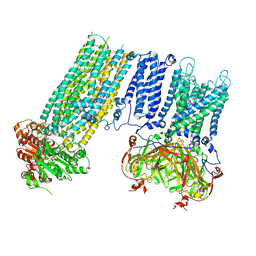 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Y
 
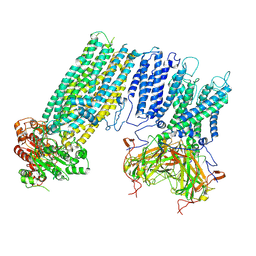 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
