8I17
 
 | |
8I03
 
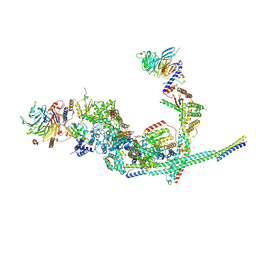 | | Cryo-EM structure of the SIN3L complex from S. pombe | | 分子名称: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | 著者 | Wang, C, Guo, Z, Zhan, X. | | 登録日 | 2023-01-10 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
8I02
 
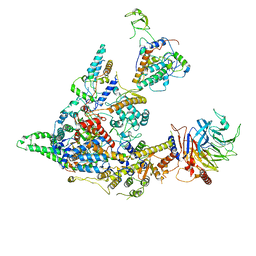 | | Cryo-EM structure of the SIN3S complex from S. pombe | | 分子名称: | Chromatin modification-related protein eaf3, Cph1, Histone deacetylase clr6, ... | | 著者 | Wang, C, Guo, Z, Zhan, X. | | 登録日 | 2023-01-10 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
8HYG
 
 | |
8HYB
 
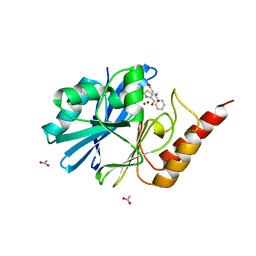 | |
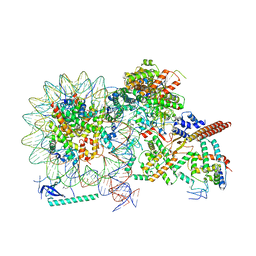
8HY0
 
 | |
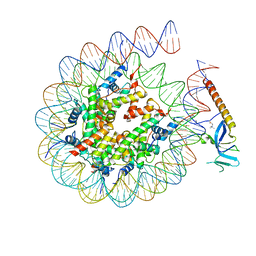
8HXZ
 
 | |
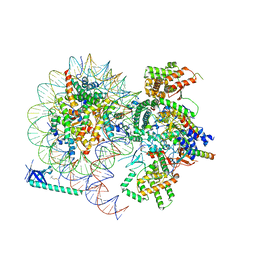
8HXY
 
 | |
8HXX
 
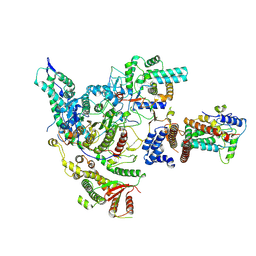 | |
8HVQ
 
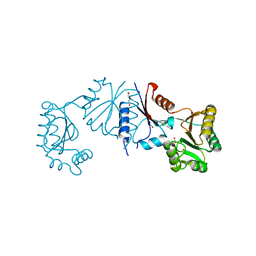 | | Crystal structure of haloacid dehalogenase-like hydrolase family enzyme from Staphylococcus lugdunensis | | 分子名称: | 1,2-ETHANEDIOL, Cof-type HAD-IIB family hydrolase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kaur, H, Mahto, J.K, Kumar, P, Sharma, A.K. | | 登録日 | 2022-12-27 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure of haloacid dehalogenase-like hydrolase family enzyme from Staphylococcus lugdunensis
To Be Published
|
|
8HRF
 
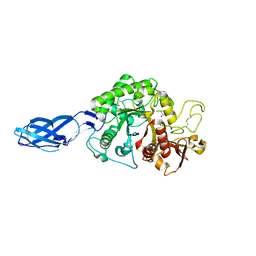 | |
8HR1
 
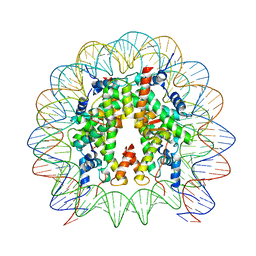 | | Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom | | 分子名称: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Zebin, T, Ai, H.S, Ziyu, X, Man, P, Liu, L. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQY
 
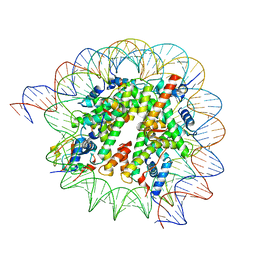 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | 分子名称: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | 著者 | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | 分子名称: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | 著者 | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | 登録日 | 2022-12-07 | | 公開日 | 2023-07-05 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8HLY
 
 | |
8HL7
 
 | |
8HKR
 
 | | Crystal Structure of Histone H3 Lysine 79 (H3K79) Methyltransferase Rv2067c from Mycobacterium tuberculosis | | 分子名称: | PHOSPHATE ION, Protein lysine methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Dadireddy, V, Singh, P.R, Kalladi, S.M, Valakunja, N, Ramakumar, S. | | 登録日 | 2022-11-28 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Mycobacterium tuberculosis methyltransferase Rv2067c manipulates host epigenetic programming to promote its own survival.
Nat Commun, 14, 2023
|
|
8HIF
 
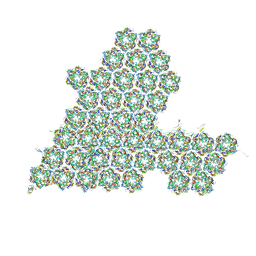 | | One asymmetric unit of Singapore grouper iridovirus capsid | | 分子名称: | Major capsid protein, Penton protein (VP14), VP137, ... | | 著者 | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | 登録日 | 2022-11-20 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
8HIA
 
 | |
8HGC
 
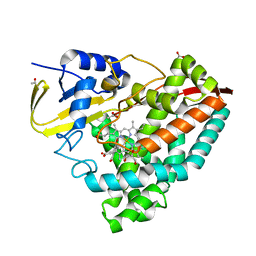 | |
8HGA
 
 | |
8HFS
 
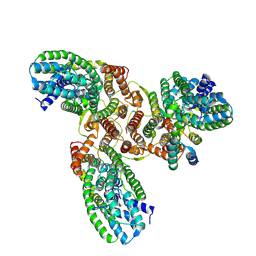 | | The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis | | 分子名称: | Bacteriocin lactococcin-A, Lactococcin-A immunity protein, Mannose-specific PTS system, ... | | 著者 | Wang, J.W. | | 登録日 | 2022-11-12 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural Basis of the Mechanisms of Action and Immunity of Lactococcin A, a Class IId Bacteriocin.
Appl.Environ.Microbiol., 89, 2023
|
|
8HEJ
 
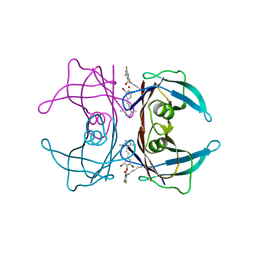 | | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole | | 分子名称: | 2,4,6-trifluorobenzaldehyde, 2,6-dibromo-4-[(E)-2-(3,5-dimethyl-1H-pyrazol-4-yl)ethenyl]phenol, Transthyretin | | 著者 | Kim, H, Choi, S, Lee, C. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole
To Be Published
|
|
8HE5
 
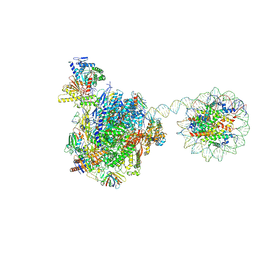 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | 分子名称: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-11-07 | | 公開日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (6.95 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
8HAN
 
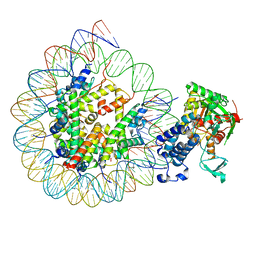 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
