9B8Z
 
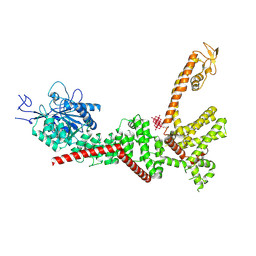 | |
9B8Y
 
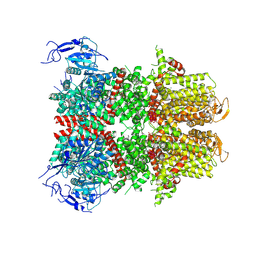 | |
9B8X
 
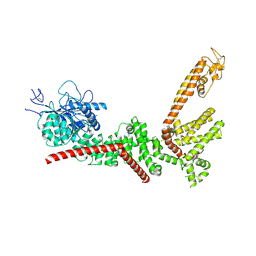 | |
9B8W
 
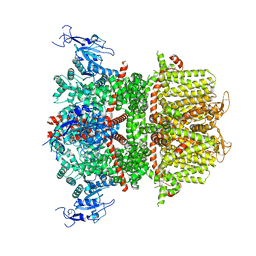 | |
9B8E
 
 | |
9B8D
 
 | |
9B7F
 
 | | S_SAD structure of HEWL using lossless default compression | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Jakoncic, J, Bernstein, H.J, Soares, A.S, Horvat, K. | | Deposit date: | 2024-03-27 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of fast and efficient lossless compression algorithms for macromolecular crystallography experiments
To Be Published
|
|
9B7E
 
 | | S_SAD structure of HEWL using lossy compression data with a compression ratio of 422 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Jakoncic, J, Bernstein, H.J, Soares, A.S, Horvat, K. | | Deposit date: | 2024-03-27 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of fast and efficient lossless compression algorithms for macromolecular crystallography experiments
To Be Published
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
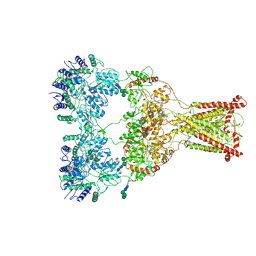
9B39
 
 | |
9B38
 
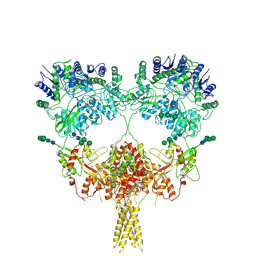 | |
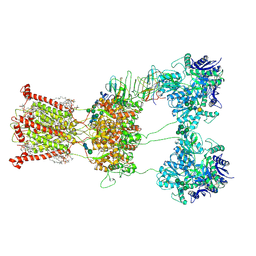
9B37
 
 | |
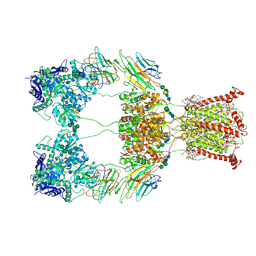
9B36
 
 | |
9B35
 
 | |
9B34
 
 | |
9B33
 
 | |
9B22
 
 | |
9B21
 
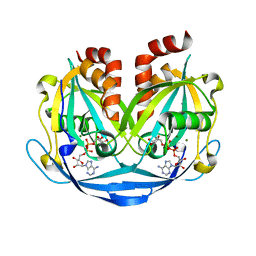 | | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form) | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-03-14 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form)
To be published
|
|
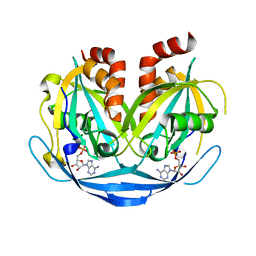
9B20
 
 | |
9B1Z
 
 | |
9B1R
 
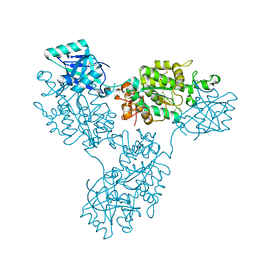 | | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vacuolar-sorting receptor 1 | | Authors: | Park, H, Youn, B, Park, D.J, Puthanveettil, S.V, Kang, C. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 (VSR1) from Arabidopsis thaliana.
Sci Rep, 14, 2024
|
|
9B0M
 
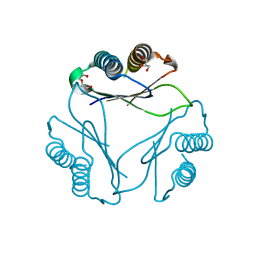 | |
9AZZ
 
 | |
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | Descriptor: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 2024
|
|
9AZI
 
 | |
