6P9K
 
 | |
6UNP
 
 | | Crystal structure of the kinase domain of BMPR2-D485G | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
5PZG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 112) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7MI5
 
 | | Asymmetrical PAM-Non PAM prespacer bound Cas4/Cas1/Cas2 complex | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (26-MER), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7MI9
 
 | | Full integration complex of Cas1/Cas2 from Cas4-containing system | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (5'-D(P*CP*GP*GP*AP*AP*AP*AP*GP*AP*GP*CP*C)-3'), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
6P9Q
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 2 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6M9G
 
 | | BbvCI B2 dimer with Ta6Br14 clusters | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BROMIDE ION, ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-23 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
8E3S
 
 | |
6TYK
 
 | |
7A9L
 
 | | Racemic compound of RNA duplexes. | | Descriptor: | RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3'), RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), ... | | Authors: | Rypniewski, W. | | Deposit date: | 2020-09-02 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Broken symmetry between RNA enantiomers in a crystal lattice.
Nucleic Acids Res., 49, 2021
|
|
6UL7
 
 | | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Gardner, S, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-10-07 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole
To be Published
|
|
7MFJ
 
 | | Structural Characterization of Beta Cyanoalanine Synthase from Tetranychus Urticae | | Descriptor: | ACETATE ION, Beta-cyanoalanine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Daneshian, L, Schlachter, C, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2021-04-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural and functional characterization of beta-cyanoalanine synthase from Tetranychus urticae.
Insect Biochem.Mol.Biol., 142, 2022
|
|
6MA3
 
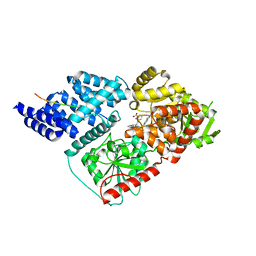 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 2a | | Descriptor: | 4-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}butanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
8DTO
 
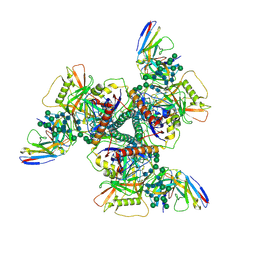 | | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-07-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
6PKG
 
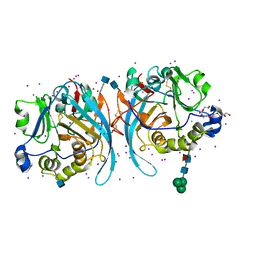 | | Zebrafish N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) catalytic domain auto-inhibited by pro-peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, IODIDE ION, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
7A9P
 
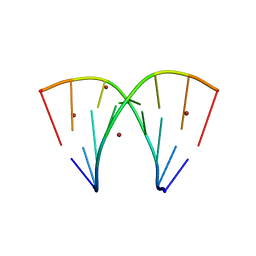 | | Racemic compound of RNA duplexes. | | Descriptor: | RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3'), RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), ... | | Authors: | Rypniewski, W. | | Deposit date: | 2020-09-02 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Broken symmetry between RNA enantiomers in a crystal lattice.
Nucleic Acids Res., 49, 2021
|
|
6TWN
 
 | | Crystal structure of Talin1 R7R8 in complex with CDK1 (206-223) | | Descriptor: | CHLORIDE ION, Cyclin-dependent kinase 1, GLYCEROL, ... | | Authors: | Zacharchenko, T, Muench, S.P, Goult, B.T. | | Deposit date: | 2020-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Talin mechanosensitivity is modulated by a direct interaction with cyclin-dependent kinase-1.
J.Biol.Chem., 297, 2021
|
|
6PKW
 
 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C2 symmetry (pseudo C4 symmetry) | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6MAC
 
 | | Ternary structure of GDF11 bound to ActRIIB-ECD and Alk5-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 11, ... | | Authors: | Goebel, E.J, Thompson, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7M2D
 
 | |
8DV4
 
 | | Crystal structure of the BC8B TCR-CD1b-PI complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | alpha beta T-cell receptor recognition of self-phosphatidylinositol presented by CD1b.
J.Biol.Chem., 299, 2023
|
|
6ZP6
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB334 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6MAL
 
 | |
6ZWU
 
 | | Racemic compound of RNA duplexes. | | Descriptor: | RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3'), RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), ... | | Authors: | Rypniewski, W. | | Deposit date: | 2020-07-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Broken symmetry between RNA enantiomers in a crystal lattice.
Nucleic Acids Res., 49, 2021
|
|
6TVK
 
 | | Alpha-L-fucosidase isoenzyme 2 from Paenibacillus thiaminolyticus | | Descriptor: | Alpha-L-fucosidase, GLYCEROL, HEXATANTALUM DODECABROMIDE, ... | | Authors: | Kovalova, T, Koval, T, Lipovova, P, Dohnalek, J. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first structure-function study of GH151 alpha-l-fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
Febs J., 2022
|
|
