8R09
 
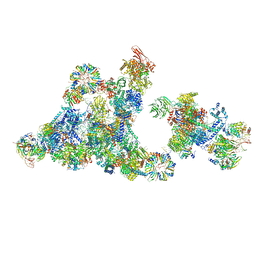 | | Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
9HGO
 
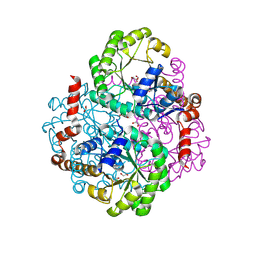 | | Crystal Structure of the Coxiella burnetii E110Q Mutant 2-methylisocitrate lyase | | Descriptor: | 1,2-ETHANEDIOL, 2-methylisocitrate lyase, CHLORIDE ION | | Authors: | Stuart, W, Isupov, M, Harmer, N.J. | | Deposit date: | 2024-11-20 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure and catalytic mechanism of methylisocitrate lyase, a potential drug target against Coxiella burnetii.
J.Biol.Chem., 301, 2025
|
|
8OJB
 
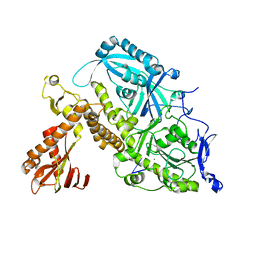 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA polymerase catalytic subunit | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
7V8K
 
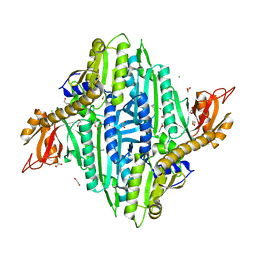 | |
5LSX
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, ZINC ION, [(2~{R},5~{S})-1-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]-5-azaniumyl-6-oxidanyl-6-oxidanylidene-hexan-2-yl]-(phenylmethyl)azanium | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
9HHY
 
 | | Crystal Structure of the Coxiella burnetii 2-methylisocitrate lyase Bound to Inhibitor Isocitric Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-methylisocitrate lyase, CHLORIDE ION, ... | | Authors: | Stuart, W, Isupov, M, Harmer, N.J. | | Deposit date: | 2024-11-22 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and catalytic mechanism of methylisocitrate lyase, a potential drug target against Coxiella burnetii.
J.Biol.Chem., 301, 2025
|
|
5ABG
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-1-[3-(4-fluorophenyl)propyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8R0A
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
9HGQ
 
 | | Crystal Structure of the Coxiella burnetii 2-methylisocitrate lyase Bound to Substrate 2-MIC | | Descriptor: | 1,2-ETHANEDIOL, 2-methylisocitrate lyase, ALPHA-METHYLISOCITRIC ACID, ... | | Authors: | Stuart, W, Isupov, M, Harmer, N.J. | | Deposit date: | 2024-11-20 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and catalytic mechanism of methylisocitrate lyase, a potential drug target against Coxiella burnetii.
J.Biol.Chem., 301, 2025
|
|
8QXD
 
 | | Cryo-EM structure of the cross-exon pre-B complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8OHO
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry H11 | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, Cyclic di-AMP synthase CdaA, MAGNESIUM ION, ... | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8OJ6
 
 | | HSV-1 DNA polymerase-processivity factor complex in pre-translocation state | | Descriptor: | DNA (22-MER), DNA (48-MER), DNA polymerase catalytic subunit, ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
3KA5
 
 | | Crystal structure of Ribosome-associated protein Y (PSrp-1) from Clostridium acetobutylicum. Northeast Structural Genomics Consortium target id CaR123A | | Descriptor: | Ribosome-associated protein Y (PSrp-1) | | Authors: | Seetharaman, J, Neely, H, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ribosome-associated protein Y (PSrp-1) from Clostridium acetobutylicum
To be Published
|
|
5O6P
 
 | |
9HRA
 
 | | Crystal Structure of the Coxiella burnetii 2-methylisocitrate lyase Bound to Products Succinic and Pyruvic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-methylisocitrate lyase, CHLORIDE ION, ... | | Authors: | Stuart, W, Isupov, M, Harmer, N.J. | | Deposit date: | 2024-12-17 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and catalytic mechanism of methylisocitrate lyase, a potential drug target against Coxiella burnetii.
J.Biol.Chem., 301, 2025
|
|
6IX7
 
 | | The structure of LepI C52A in complex with SAH and substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
8OJA
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
5FL0
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d] [1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
8OJD
 
 | | HSV-1 DNA polymerase beta-hairpin loop | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
5O11
 
 | |
8YMI
 
 | | BRD4-BD1 in complex with 2-{[(3S)-1-benzylpyrrolidin-3-yl]methyl}-5-methyl-7-(1-methylpyrazol-3-yl)pyrazolo[4,3-c]pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(3S)-1-benzylpyrrolidin-3-yl]methyl}-5-methyl-7-(1-methylpyrazol-3-yl)pyrazolo[4,3-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Discovery of a potent, orally available pyrazolopyridone derivative as a novel selective BET BD1 Inhibitor (Part 2).
To Be Published
|
|
6EIJ
 
 | | DYRK1A in complex with HG-8-60-1 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}-[5-[3-(2-morpholin-4-ylethylcarbamoylamino)phenyl]-[1,3]thiazolo[5,4-b]pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7NH8
 
 | | Crystal structure of human carbonic anhydrase II with N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Synthesis of Azasugar-Sulfonamide conjugates and their Evaluation as Inhibitors of Carbonic Anhydrases: the Azasugar Approach to Selectivity
Eur.J.Org.Chem., 2021
|
|
6EMN
 
 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
1G95
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE GLMU, APO FORM | | Descriptor: | N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Kostrewa, D, D'Arcy, A, Kamber, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
J.Mol.Biol., 305, 2001
|
|
