8TSY
 
 | |
8AKH
 
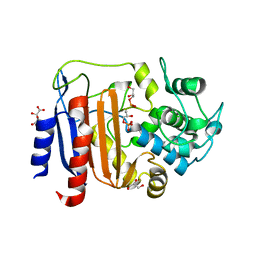 | | Crystal structure of DltE from L. plantarum soaked with LTA | | Descriptor: | Beta-lactamase family protein, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
6JGE
 
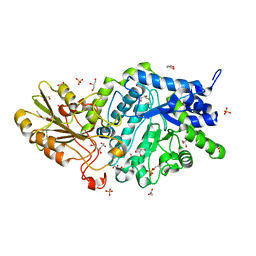 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
7AKQ
 
 | |
8AGR
 
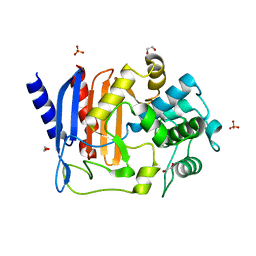 | | Crystal structure of DltE from L. plantarum, apo form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase family protein, SULFATE ION | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
7REG
 
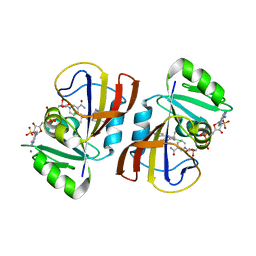 | | DfrA1 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, CALCIUM ION, Dihydrofolate reductase type 1, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
8T8L
 
 | |
7AL9
 
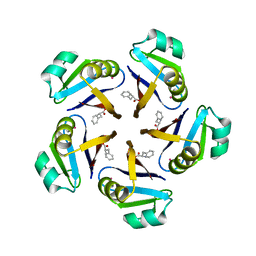 | |
7ALA
 
 | | human GCH-GFRP inhibitory complex | | Descriptor: | 2-azanyl-8-[(4-fluorophenyl)methylsulfanyl]-1,7-dihydropurin-6-one, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8AV0
 
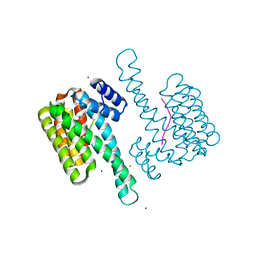 | | small molecule stabilizer (compound 1) for C-RAF pS259 and 14-3-3 | | Descriptor: | 1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-4-carboxamide, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
7RK4
 
 | | Mannitol-2-dehydrogenase from Aspergillus fumigatus | | Descriptor: | Mannitol 2-dehydrogenase | | Authors: | Nguyen, S, Bruning, J.B. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the Mannitol Biosynthesis Pathway in Aspergillus fumigatus: Characterisation and Inhibition of Mannitol-2-Dehydrogenase
To Be Published
|
|
6CSP
 
 | |
7ALC
 
 | | human GCH-GFRP stimulatory complex | | Descriptor: | GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, PHENYLALANINE, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6JG9
 
 | |
7R27
 
 | | Crystal structure of the L. plantarum D-alanine ligase DltA | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-ALANINE, D-alanine--D-alanyl carrier protein ligase | | Authors: | Nikolopoulos, N, Ravaud, S, Simorre, J.P, Grangeasse, C. | | Deposit date: | 2022-02-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | DltC acts as an interaction hub for AcpS, DltA and DltB in the teichoic acid D-alanylation pathway of Lactiplantibacillus plantarum.
Sci Rep, 12, 2022
|
|
6CQC
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
7ALY
 
 | |
8TSZ
 
 | |
8AJI
 
 | | Crystal structure of DltE from L. plantarum, TCEP form | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Beta-lactamase family protein, GLYCEROL, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
7ALB
 
 | | human GCH-GFRP stimulatory complex 7-deaza-GTP bound | | Descriptor: | 7-deaza-GTP, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6JGN
 
 | | Crystal structure of barley exohydrolaseI W434H in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
7QZR
 
 | |
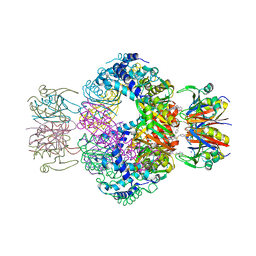
7AWP
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with rubidium and barium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AMX
 
 | |
