9BS4
 
 | | DNA Ligase 1 E346A/E592A double mutant with 5'-rG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | KanalElamparithi, B, Caglayan, M. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
J.Biol.Chem., 300, 2024
|
|
8BA6
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
6T2W
 
 | | Crystal structure of the CSF1R kinase domain with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
7ZEI
 
 | | Thermostable GH159 glycoside hydrolase from Caldicellulosiruptor at 1.7 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Baudrexl, M, Fida, T, Berk, B, Schwarz, W, Zverlov, V.V, Groll, M, Liebl, W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Thermostable GH159 Glycoside Hydrolases Exhibiting alpha-L-Arabinofuranosidase Activity.
Front Mol Biosci, 9, 2022
|
|
8YAZ
 
 | | XFEL crystal structure of the oxidized form of F87A/F393H P450BM3 with N-enanthyl-L-prolyl-L-phenylalanine in complex with styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Nagao, S, Kuwano, W, Tosha, T, Yamashita, K, Stanfield, J.K, Kasai, C, Ariyasu, S, Shoji, O, Sugimoto, H, Kubo, M. | | Deposit date: | 2024-02-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3.
Commun Chem, 8, 2025
|
|
7LJI
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Gd+3 bound | | Descriptor: | GADOLINIUM ION, Poly(Aspartic acid) hydrolase | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
7LJH
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Zn+2 bound | | Descriptor: | Poly(Aspartic acid) hydrolase, ZINC ION | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
5JOU
 
 | | Bacteroides ovatus Xyloglucan PUL GH31 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase BoGH31A, NICKEL (II) ION | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
1FCH
 
 | | CRYSTAL STRUCTURE OF THE PTS1 COMPLEXED TO THE TPR REGION OF HUMAN PEX5 | | Descriptor: | PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR, PTS1-CONTAINING PEPTIDE | | Authors: | Gatto Jr, G.J, Geisbrecht, B.V, Gould, S.J, Berg, J.M. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5.
Nat.Struct.Biol., 7, 2000
|
|
6LT7
 
 | |
8EXG
 
 | | Human Carbonic Anhydrase II bound N-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)-4-sulfamoylbenzamide | | Descriptor: | (4S)-5-amino-4-{1,3-dioxo-4-[(2-{2-[2-(4-sulfamoylbenzamido)ethoxy]ethoxy}ethyl)amino]-1,3-dihydro-2H-isoindol-2-yl}-5-oxopentanoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8H6K
 
 | | Cryo-EM structure of human exon-defined spliceosome in the mature B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8YB1
 
 | | XFEL crystal structure of the reduced form of F87A/F393H P450BM3 with N-enanthyl-L-prolyl-L-phenylalanine in complex with styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Nagao, S, Kuwano, W, Tosha, T, Yamashita, K, Stanfield, J.K, Kasai, C, Ariyasu, S, Shoji, O, Sugimoto, H, Kubo, M. | | Deposit date: | 2024-02-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3.
Commun Chem, 8, 2025
|
|
7SS7
 
 | | Crystal structure of Klebsiella LpxH in complex with JH-LPH-50 | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(4-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}phenyl)carbamoyl]amino}-N-hydroxyhexanamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
8YB2
 
 | | XFEL crystal structure of the oxygen-bound form of F393H P450BM3 with N-enanthyl-L-prolyl-L-phenylalanine in complex with styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Nagao, S, Kuwano, W, Tosha, T, Yamashita, K, Stanfield, J.K, Kasai, C, Ariyasu, S, Shoji, O, Sugimoto, H, Kubo, M. | | Deposit date: | 2024-02-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3.
Commun Chem, 8, 2025
|
|
8YB3
 
 | | XFEL crystal structure of the oxygen-bound form of F87A/F393H P450BM3 with N-enanthyl-L-prolyl-L-phenylalanine in complex with styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Nagao, S, Kuwano, W, Tosha, T, Yamashita, K, Stanfield, J.K, Kasai, C, Ariyasu, S, Shoji, O, Sugimoto, H, Kubo, M. | | Deposit date: | 2024-02-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3.
Commun Chem, 8, 2025
|
|
7FZ3
 
 | | Crystal Structure of human FABP4 in complex with 4-(4-chlorophenoxy)benzenesulfinic acid:sodium hydride, i.e. SMILES c1(Oc2ccc(cc2)Cl)ccc(cc1)[S@@](=O)O with IC50=7.4 microM | | Descriptor: | 1-[bis(oxidanyl)-$l^{3}-sulfanyl]-4-(4-chloranylphenoxy)benzene, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6S30
 
 | | Crystal Structure of Seryl-tRNA Synthetase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine--tRNA ligase | | Authors: | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
8YAY
 
 | | XFEL crystal structure of the oxidized form of F393H P450BM3 with N-enanthyl-L-prolyl-L-phenylalanine in complex with styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Nagao, S, Kuwano, W, Tosha, T, Yamashita, K, Stanfield, J.K, Kasai, C, Ariyasu, S, Shoji, O, Sugimoto, H, Kubo, M. | | Deposit date: | 2024-02-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3.
Commun Chem, 8, 2025
|
|
8KAE
 
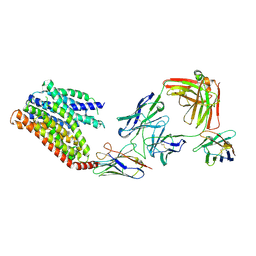 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
8H6J
 
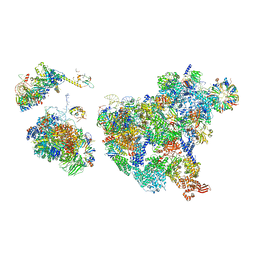 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8B5G
 
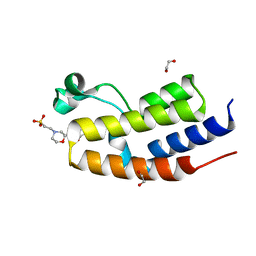 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-3-methyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,8-dimethoxy-3-methyl-1~{H}-3-benzazepin-2-one, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8BA5
 
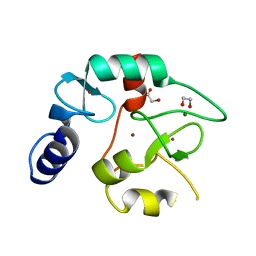 | | Crystal structure of the DNMT3A ADD domain | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, MAGNESIUM ION, ... | | Authors: | Linhard, V, Kunert, S, Wollenhaupt, J, Broehm, A, Schwalbe, H, Jeltsch, A. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The MECP2-TRD domain interacts with the DNMT3A-ADD domain at the H3-tail binding site.
Protein Sci., 32, 2023
|
|
8H6L
 
 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
4UIT
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 7-(3,4-dimethoxyphenyl)-2-(4- methanesulfonylpiperazine-1-carbonyl)-5-methyl-4H,5H-thieno-3,2-c- pyridin-4-one | | Descriptor: | 7-(3,4-dimethoxyphenyl)-5-methyl-2-(4-methylsulfonylpiperazin-1-yl)carbonyl-thieno[3,2-c]pyridin-4-one, BROMODOMAIN-CONTAINING PROTEIN 9 | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
