5HDB
 
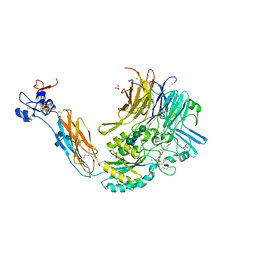 | | Integrin alphaIIbbeta3 in complex with Ro-435054 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lin, F.Y. | | Deposit date: | 2016-01-05 | | Release date: | 2016-02-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7012 Å) | | Cite: | beta-Subunit Binding Is Sufficient for Ligands to Open the Integrin alpha IIb beta 3 Headpiece.
J.Biol.Chem., 291, 2016
|
|
8OXO
 
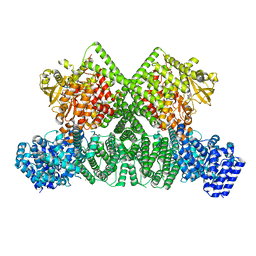 | | ATM(Q2971A) dimeric C-terminal region activated by oxidative stress in complex with Mg AMP-PNP and p53 peptide | | Descriptor: | Cellular tumor antigen p53, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
8P2D
 
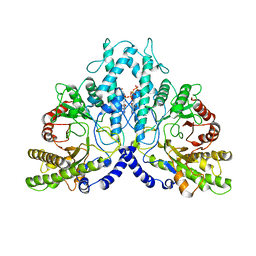 | | Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P39
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-17 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To be published
|
|
8P1H
 
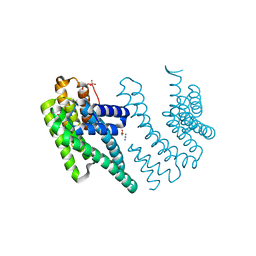 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
8PBB
 
 | | CHAPSO treated partial catalytic component (comprising only AnfD & AnfK, lacking AnfG and FeFeco) of iron nitrogenase from Rhodobacter capsulatus | | Descriptor: | FE(8)-S(7) CLUSTER, Nitrogenase iron-iron protein, beta subunit, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PCW
 
 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8P24
 
 | |
8P1B
 
 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1C
 
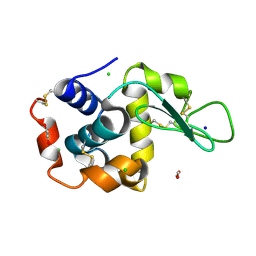 | | Lysozyme structure solved from serial crystallography data collected at 1 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1A
 
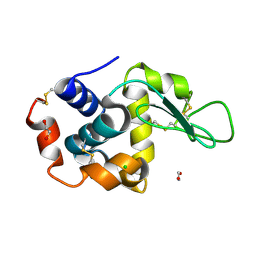 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz for 5 seconds with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8OJN
 
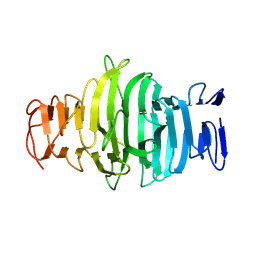 | |
8OML
 
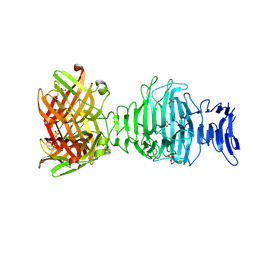 | |
8ONC
 
 | |
8P1D
 
 | | Lysozyme structure solved from serial crystallography data collected at 100 Hz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8ONF
 
 | |
8OK3
 
 | |
8OKH
 
 | |
8PBX
 
 | |
8OKS
 
 | |
8ON4
 
 | |
8ONB
 
 | |
1R80
 
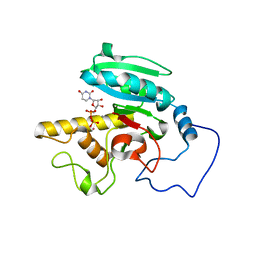 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
8OND
 
 | |
8OOH
 
 | |
