8QQI
 
 | |
6E8G
 
 | |
6OKW
 
 | | Solution structure of VEK50 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6I7C
 
 | | Dye type peroxidase Aa from Streptomyces lividans: imidazole complex | | Descriptor: | Deferrochelatase/peroxidase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno-Chicano, T, Ebrahim, A.E, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6OQ9
 
 | |
6OQP
 
 | | U-AITx-Ate1 | | Descriptor: | SER-LYS-TRP-ILE-CYS-ALA-ASN-ARG-SER-VAL-CYS-PRO-ILE | | Authors: | Elnahriry, K.A, Wai, D.C.C, Norton, R.S. | | Deposit date: | 2019-04-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterisation of a novel peptide from the Australian sea anemone Actinia tenebrosa.
Toxicon, 168, 2019
|
|
6OSW
 
 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
6OKY
 
 | | Solution structure of truncated peptide from PAMap53 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6P5W
 
 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6P6B
 
 | |
8S9S
 
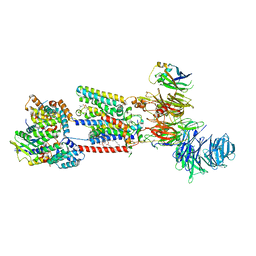 | | Structure of the human ER membrane protein complex (EMC) in GDN | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tomaleri, G.P, Nguyen, V.N, Voorhees, R.M. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A selectivity filter in the ER membrane protein complex limits protein misinsertion at the ER.
J.Cell Biol., 222, 2023
|
|
5XG7
 
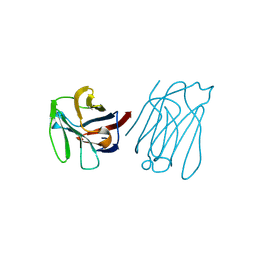 | | Galectin-13/Placental Protein 13 crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y, Wang, Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
6P6C
 
 | |
6EZN
 
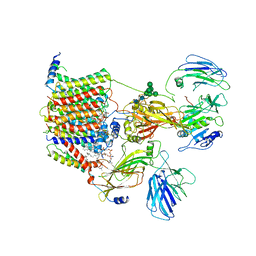 | | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Kowal, J, Eyring, J, Ngwa, E.M, Aebi, M, Locher, K.P. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-17 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
6P9L
 
 | |
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5XF4
 
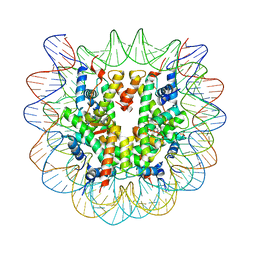 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (S,S-configuration) | | Descriptor: | (1S,2S)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
6P9K
 
 | |
5XQM
 
 | |
5XS1
 
 | |
6PGE
 
 | |
5X7X
 
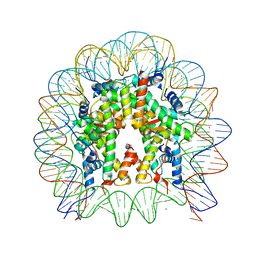 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
8PK7
 
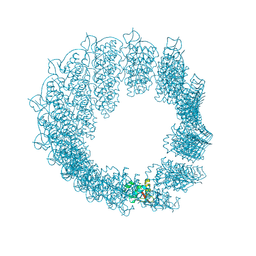 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The alphavirus nsP3 protein forms helical tubular scaffolds important for viral replication and particle assembly
To Be Published
|
|
8PHZ
 
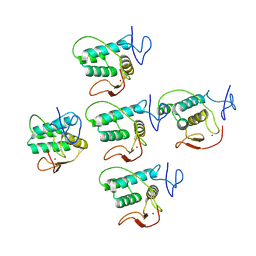 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The alphavirus nsP3 protein forms helical tubular scaffolds important for viral replication and particle assembly
To be published
|
|
8QE8
 
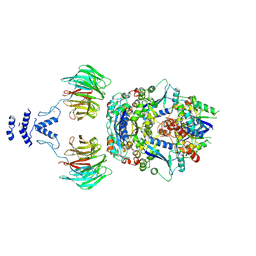 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
