1ZPI
 
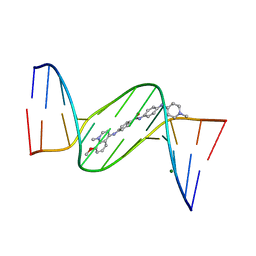 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8224 complexed with CGCGAATTCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', 8-METHOXY-1-METHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1EEG
 
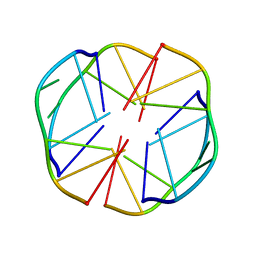 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
1RPF
 
 | |
2DES
 
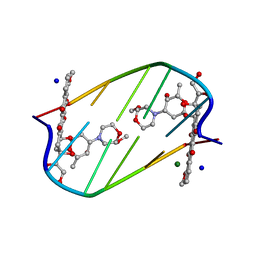 | | INTERACTIONS BETWEEN MORPHOLINYL ANTHRACYCLINES AND DNA: THE CRYSTAL STRUCTURE OF A MORPHOLINO DOXORUBICIN BOUND TO D(CGTACG) | | Descriptor: | 3'-DESAMINO-3'-(2-METHOXY-4-MORPHOLINYL)-DOXORUBICIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Cirilli, M, Bachechi, F, Ughetto, G, Colonna, F.P, Capobianco, M.L. | | Deposit date: | 1993-03-16 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions between morpholinyl anthracyclines and DNA. The crystal structure of a morpholino doxorubicin bound to d(CGTACG).
J.Mol.Biol., 230, 1993
|
|
7ESN
 
 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, H105F Rha-GlcA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, L-Rhamnose-alpha-1,4-D-glucuronate lyase, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
2VVP
 
 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with its substrates ribose 5-phosphate and ribulose 5-phosphate | | Descriptor: | 5-O-phosphono-D-ribose, RIBOSE-5-PHOSPHATE ISOMERASE B, RIBULOSE-5-PHOSPHATE | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
5XF2
 
 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1U40
 
 | | IspF with 4-diphosphocytidyl-2C-methyl-D-erythritol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
2ZTU
 
 | | T190A mutant of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
3ITA
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in acyl-enzyme complex with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase dacC, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
1U43
 
 | | IspF with 4-diphosphocytidyl-2c-methyl-D-erythritol 2-phosphate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL 2-PHOSPHATE | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1G80
 
 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
4MOL
 
 | | Pyranose 2-oxidase H167A mutant with 2-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-galactopyranose, DODECAETHYLENE GLYCOL, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
3A3Z
 
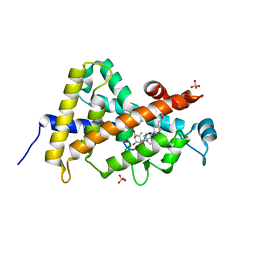 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277A(C23S) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-2-methyl-9,10-secoandrosta-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
4MIH
 
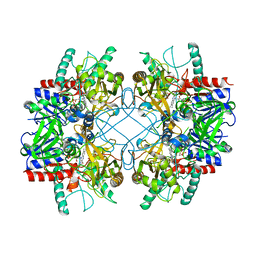 | | Pyranose 2-oxidase from Phanerochaete chrysosporium, recombinant H158A mutant | | Descriptor: | 3-deoxy-3-fluoro-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Hassan, N, Tan, T.C, Spadiut, O, Pisanelli, I, Fusco, L, Haltrich, D, Peterbauer, C, Divne, C. | | Deposit date: | 2013-08-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Phanerochaete chrysosporium pyranose 2-oxidase suggest that the N-terminus acts as a propeptide that assists in homotetramer assembly.
FEBS Open Bio, 3, 2013
|
|
5OCE
 
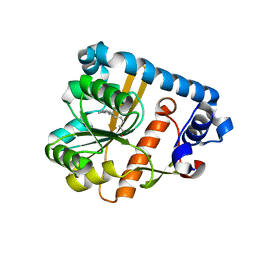 | | THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF THE MEMBRANE ACYLTRANSFERASE PatA -- Complex of PatA with palmitate, mannose, and palmitoyl-6-mannose | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase, [(2~{R},3~{S},4~{S},5~{S},6~{S})-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hexadecanoate, ... | | Authors: | Albesa-Jove, D, Tersa, M, Guerin, M.E. | | Deposit date: | 2017-06-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Molecular Mechanism of Substrate Recognition and Catalysis of the Membrane Acyltransferase PatA from Mycobacteria.
ACS Chem. Biol., 13, 2018
|
|
3A2H
 
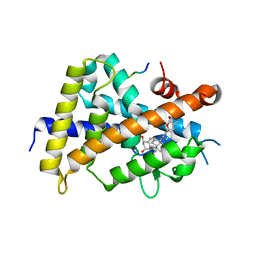 | | Crystal structure of the rat vitamin D receptor ligand binding domain complexed with TEI-9647 and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | (1S,3R,5Z,7E,20S,23S)-1,3-dihydroxy-23,26-epoxy-9,10-secocholesta-5,7,10,25(27)-tetraen-26-one, Mediator of RNA polymerase II transcription subunit 1 peptide, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2009-05-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the histidine-mediated vitamin D receptor agonistic and antagonistic mechanisms of (23S)-25-dehydro-1alpha-hydroxyvitamin D(3)-26,23-lactone
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5OWY
 
 | | Glycogen Phosphorylase in complex with KS252 | | Descriptor: | 4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G, Kantsadi, A, Chatzileontiadou, D, Leonidas, D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
5OX1
 
 | | Glycogen Phosphorylase in complex with JLH270 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-methoxyphenyl)-1~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
5O56
 
 | | Glycogen Phosphorylase b in complex with 29b | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | Authors: | Solovou, T.G.A, Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
2EKQ
 
 | |
3L1V
 
 | | Crystal structure of GmhB from E. coli in complex with calcium and phosphate. | | Descriptor: | CALCIUM ION, D,D-heptose 1,7-bisphosphate phosphatase, PHOSPHATE ION, ... | | Authors: | Sugiman-Marangos, S.N, Junop, M.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
6IBR
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclophellitol epoxide LWA481 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-5-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
6BBE
 
 | | Structure of N-glycosylated porcine surfactant protein-D | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van Eijk, M, Rynkiewicz, M.J, Khatri, K, Leymarie, N, Zaia, J, White, M.R, Hartshorn, K.L, Cafarella, T.R, van Die, I, Hessing, M, Seaton, B.A, Haagsman, H.P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-05-23 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus.
J. Biol. Chem., 293, 2018
|
|
4YTS
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy 3-keto D-galactitol | | Descriptor: | 1-deoxy-D-xylo-hex-3-ulose, 1-deoxy-alpha-D-xylo-hex-3-ulofuranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
