108D
 
 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
204D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
203D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
1G80
 
 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
1G7Z
 
 | | NMR SOLUTION STRUCTURE OF D(CGCTAGCG)2 | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
4RNQ
 
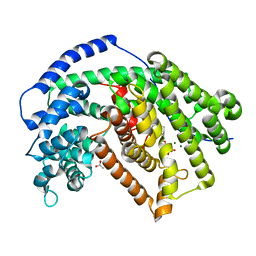 | | Crystal structure of tobacco 5-epi-aristolochene synthase (TEAS) with anilinogeranyl diphosphate (AGPP) and geraniline | | Descriptor: | (2E,6E)-3,7-dimethyl-8-(phenylamino)octa-2,6-dien-1-yl trihydrogen diphosphate, 5-epi-aristolochene synthase, ACETATE ION, ... | | Authors: | Koo, H.J, Crenshaw, C.M, Starks, C, Spielmann, H.P, Chappell, J, Noel, J.P. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Formation of a Novel Macrocyclic Alkaloid from the Unnatural Farnesyl Diphosphate Analogue Anilinogeranyl Diphosphate by 5-Epi-Aristolochene Synthase.
Acs Chem.Biol., 10, 2015
|
|
1KKW
 
 | |
1KKV
 
 | |
214D
 
 | | THE SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING A SINGLE 2'-O-METHYL-BETA-D-ARAT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*AP*TP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*AP*(T41)P*AP*TP*GP*CP*G)-3') | | Authors: | Gotfredsen, C.H, Spielmann, H.P, Wengel, J, Jacobsen, J.P. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA duplex containing a single 2'-O-methyl-beta-D-araT: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Bioconjug.Chem., 7, 1996
|
|
