3SQ4
 
 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
6JEZ
 
 | | Covalent labeling of rVDR-LBD by turn-on fluorescent probe mediated by conjugate addition and cyclization | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, 7-(diethylamino)chromen-2-one, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
7VQP
 
 | | Vitamin D receptor complexed with a lithocholic acid derivative | | Descriptor: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
7EWF
 
 | |
7EWM
 
 | |
1HEI
 
 | |
2M14
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and Rad4 | | Descriptor: | DNA repair protein RAD4, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lafrance-Vanasse, J, Arseneault, G, Cappadocia, L, Legault, P, Omichinski, J.G. | | Deposit date: | 2012-11-16 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER.
Nucleic Acids Res., 41, 2013
|
|
6FDP
 
 | |
8OQI
 
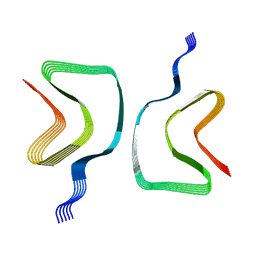 | | Cryo-EM structure of the wild-type alpha-synuclein fibril. | | Descriptor: | Alpha-synuclein | | Authors: | Pesch, V, Reithofer, S, Ma, L, Flores-Fernandez, J.M, Oezduezenciler, P, Busch, Y, Lien, Y, Rudtke, O, Frieg, B, Schroeder, G.F, Wille, H, Tamgueney, G. | | Deposit date: | 2023-04-12 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Vaccination with structurally adapted fungal protein fibrils induces immunity to Parkinson's disease.
Brain, 147, 2024
|
|
6FDT
 
 | |
1HUO
 
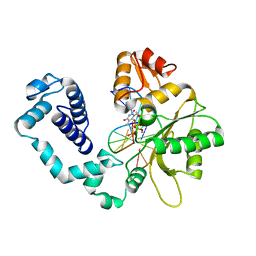 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1HUZ
 
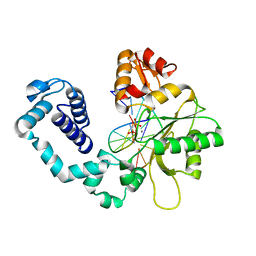 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
3FTM
 
 | |
2LOX
 
 | |
8V7F
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T*())-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+
To Be Published
|
|
7AQK
 
 | | Model of the actin filament Arp2/3 complex branch junction in cells | | Descriptor: | Actin, alpha skeletal muscle, ACTA1, ... | | Authors: | Faessler, F, Dimchev, G, Hodirnau, V.V, Wan, W, Schur, F.K.M. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction.
Nat Commun, 11, 2020
|
|
5JW9
 
 | |
4A8J
 
 | | Crystal Structure of the Elongator subcomplex Elp456 | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Elongator Subcomplex Elp456 is a Hexameric Reca-Like ATPase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3FS0
 
 | |
6ALY
 
 | | Solution structure of yeast Med15 ABD2 residues 277-368 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Tuttle, L.M, Pacheco, D, Warfield, L, Hahn, S, Klevit, R.E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex.
Cell Rep, 22, 2018
|
|
7XR3
 
 | |
7XR2
 
 | |
2ZMI
 
 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]pent-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2DB9
 
 | | Solution structure of the plus-3 domain of human KIAA0252 protein | | Descriptor: | Paf1/RNA polymerase II complex component | | Authors: | Yoneyama, M, Kigawa, T, Tochio, N, Nameki, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-06-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the plus-3 domain of human KIAA0252 protein
To be Published
|
|
