6SVL
 
 | |
1BYL
 
 | |
5ME7
 
 | | Crystal Structure of eiF4E from C. melo | | Descriptor: | Eukaryotic transcription initiation factor 4E, GLYCEROL | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
6SSH
 
 | | Structure of the TSC2 GAP domain | | Descriptor: | 1,2-ETHANEDIOL, GTPase activator-like protein | | Authors: | Hansmann, P, Kiontke, S, Kummel, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the TSC2 GAP Domain: Mechanistic Insight into Catalysis and Pathogenic Mutations.
Structure, 28, 2020
|
|
2HAP
 
 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
7K66
 
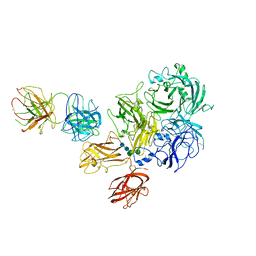 | | Structure of Blood Coagulation Factor VIII in Complex with an Anti-C1 Domain Pathogenic Antibody Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A9 heavy chain, ... | | Authors: | Childers, K.C, Gish, J, Jarvis, L, Peters, S, Garrels, C, Smith, I.W, Spencer, H.T, Spiegel, P.C. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of blood coagulation factor VIII in complex with an anti-C1 domain pathogenic antibody inhibitor.
Blood, 137, 2021
|
|
8FNB
 
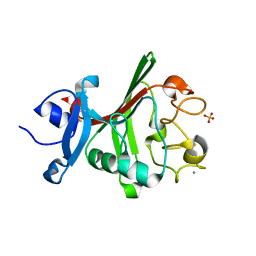 | |
2HAC
 
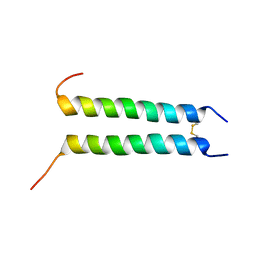 | | Structure of Zeta-Zeta Transmembrane Dimer | | Descriptor: | T-cell surface glycoprotein CD3 zeta chain | | Authors: | Chou, J.J, Wucherpfennig, K.W, Schnell, J.R, Call, M.E. | | Deposit date: | 2006-06-12 | | Release date: | 2006-10-31 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
6NOU
 
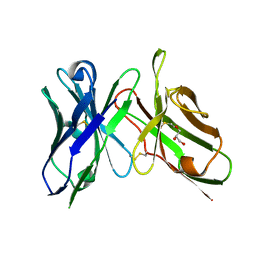 | | An scFv derived from ixekizumab | | Descriptor: | GLYCEROL, scFv derived from ixekizumab | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
6SW6
 
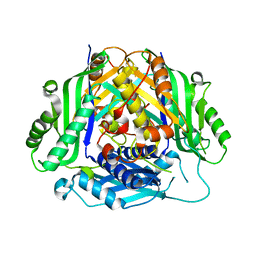 | |
8FNA
 
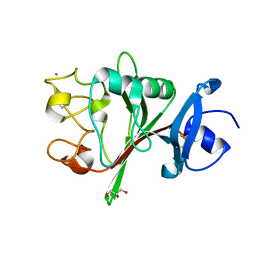 | | Crystal structure of the C-terminal Fg domain of human TNR | | Descriptor: | CALCIUM ION, GLYCEROL, Tenascin-R | | Authors: | Bouyain, S. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein-protein interactions between tenascin-R and RPTP zeta /phosphacan are critical to maintain the architecture of perineuronal nets.
J.Biol.Chem., 299, 2023
|
|
6SD5
 
 | | Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6O06
 
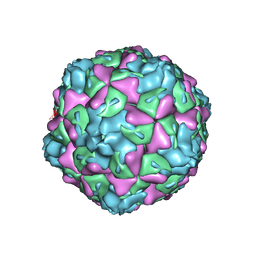 | | Extracellular factors prime enterovirus particles for uncoating | | Descriptor: | VP1, VP2, VP3 | | Authors: | Domanska, A, Ruokolainen, V, Pelliccia, M, Laajala, M, Butcher, S.J, Marjomaki, V.S. | | Deposit date: | 2019-02-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Extracellular Albumin and Endosomal Ions Prime Enterovirus Particles for Uncoating That Can Be Prevented by Fatty Acid Saturation.
J.Virol., 93, 2019
|
|
6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8SMV
 
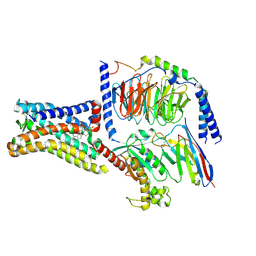 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
5KDF
 
 | | Inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with inhibitor 6 and inorganic pyrophosphate | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, PYROPHOSPHATE 2-, ... | | Authors: | Pang, A.H, Garzan, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis.
ACS Chem. Biol., 11, 2016
|
|
6C26
 
 | | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex | | Descriptor: | (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structure of a eukaryotic oligosaccharyltransferase complex.
Nature, 555, 2018
|
|
5MRA
 
 | | human SCBD (sorcin calcium binding domain) in complex with doxorubicin | | Descriptor: | DIMETHYL SULFOXIDE, DOXORUBICIN, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Colotti, G, Genovese, I. | | Deposit date: | 2016-12-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Binding of doxorubicin to Sorcin impairs cell death and increases drug resistance in cancer cells.
Cell Death Dis, 8, 2017
|
|
1BND
 
 | | STRUCTURE OF THE BRAIN-DERIVED NEUROTROPHIC FACTOR(SLASH)NEUROTROPHIN 3 HETERODIMER | | Descriptor: | BRAIN DERIVED NEUROTROPHIC FACTOR, ISOPROPYL ALCOHOL, NEUROTROPHIN 3 | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1996-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer.
Biochemistry, 34, 1995
|
|
5NBW
 
 | |
1G2K
 
 | | HIV-1 PROTEASE WITH CYCLIC SULFAMIDE INHIBITOR, AHA047 | | Descriptor: | 3-(7-BENZYL-4,5-DIHYDROXY-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1L6-[1,2,7]THIADIAZEPAN-2-YLMETHYL)-N-METHYL-BENZAMIDE, PROTEASE RETROPEPSIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-20 | | Release date: | 2001-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
8FN8
 
 | |
6SPK
 
 | | A4V MUTANT OF HUMAN SOD1 WITH EBSELEN DERIVATIVE 6 | | Descriptor: | 2-selanyl-~{N}-[3-[4-(trifluoromethyl)phenyl]phenyl]benzamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Chantadul, V, Amporndanai, K, Wright, G, Shahid, M, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-09-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
5KGX
 
 | | HIV1 catalytic core domain in complex with an inhibitor (2~{S})-2-[3-(3,4-dihydro-2~{H}-chromen-6-yl)-1-methyl-indol-2-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-methyl-1H-indol-2-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Feng, L, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-10-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Indole-based allosteric inhibitors of HIV-1 integrase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6C8W
 
 | |
