7ZMH
 
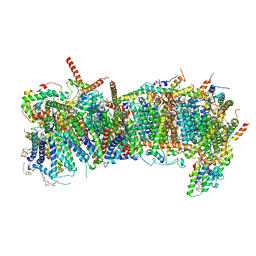 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
8OHA
 
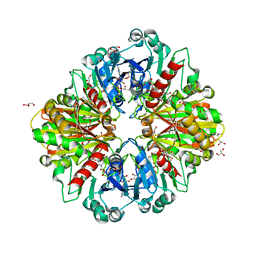 | | Crystal structure of Leptospira interrogans GAPDH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Navas-Yuste, S, de la Paz, K, Querol-Garcia, J, Gomez-Quevedo, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity.
Front Immunol, 14, 2023
|
|
8A49
 
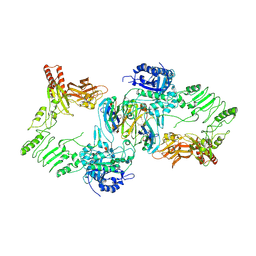 | | Endoglycosidase S in complex with IgG1 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, Secreted endoglycosidase EndoS | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
7JPK
 
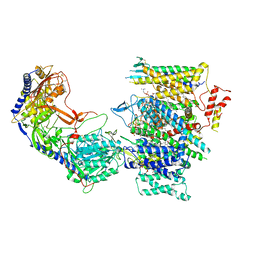 | | Rabbit Cav1.1 in the presence of 100 micromolar (S)-(-)-Bay K8644 in nanodiscs at 3.0 Angstrom resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, ... | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-08-09 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5T04
 
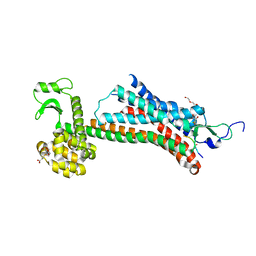 | | STRUCTURE OF CONSTITUTIVELY ACTIVE NEUROTENSIN RECEPTOR | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ARG-ARG-PRO-TYR-ILE-LEU, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B, Botos, I, Grisshammer, R. | | Deposit date: | 2016-08-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and dynamics of a constitutively active neurotensin receptor.
Sci Rep, 6, 2016
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
7ZM7
 
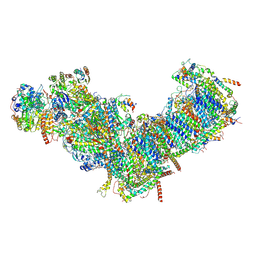 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7K4X
 
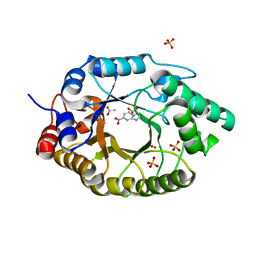 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
8OIY
 
 | |
6VHD
 
 | |
5T3P
 
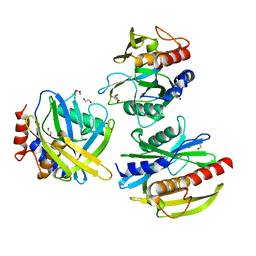 | | Crystal structure of Human Peroxisomal coenzyme A diphosphatase NUDT7 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal coenzyme A diphosphatase NUDT7 | | Authors: | Srikannathasan, V, Nunez, C.A, Tallant, C, Siejka, P, Mathea, S, Kopec, J, Elkins, J.M, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Human Peroxisomal coenzyme A diphosphatase NUDT7
To Be Published
|
|
3OF0
 
 | |
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5YQA
 
 | | The crystal structure of CYP199A4 binding with 4-n-Propyl benzoic acid | | Descriptor: | 4-propylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Zhang, T, Qiao, R, Bell, S, Coleman, T. | | Deposit date: | 2017-11-06 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The crystal structure of CYP199A4 binding with 4-n-Propyl benzoic acid
To Be Published
|
|
8QJZ
 
 | | Crystal structure of E. coli LpxH in complex with lipid X | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5AJR
 
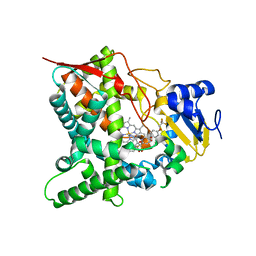 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma cruzi in complex with the 1-tetrazole derivative VT-1161 ((R)-2-(2,4-Difluorophenyl)-1, 1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl) pyridin-2-yl)propan-2-ol) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Wawrzak, Z, Hoekstra, W, I Lepesheva, G. | | Deposit date: | 2015-02-26 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Clinical Candidate Vt-1161'S Antiparasitic Effect in Vitro, Activity in a Murine Model of Chagas Disease, and Structural Characterization in Complex with the Target Enzyme Cyp51 from Trypanosoma Cruzi.
Antimicrob.Agents Chemother., 60, 2015
|
|
6V3R
 
 | | Crystal structure of murine cycloxygenase in complex with a harmaline analog, 4,9-dihydro-3H-pyrido[3,4-b]indole | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-[(4-chlorophenyl)methyl]-6-methoxy-1-methyl-4,9-dihydro-3H-beta-carboline, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Harmaline Analogs as Substrate-Selective Cyclooxygenase-2 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7ZHZ
 
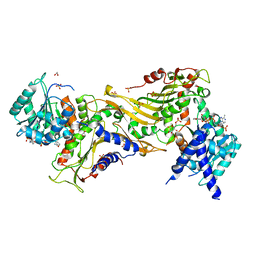 | |
5AK9
 
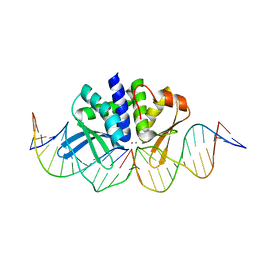 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3, ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
8QKA
 
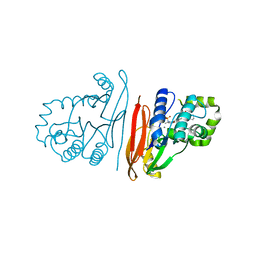 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5AKM
 
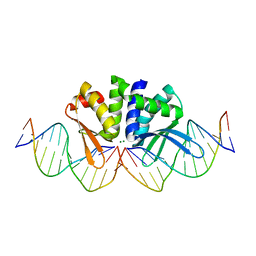 | | THE CRYSTAL STRUCTURE OF I-DMOI G20S IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MG | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5TVV
 
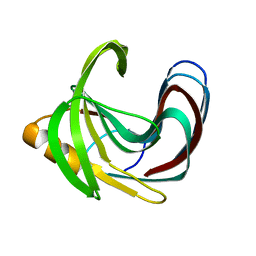 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | Descriptor: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
5YSW
 
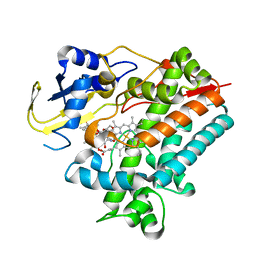 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5LTW
 
 | | Complex of human 14-3-3 sigma CLU1 mutant with phosphorylated heat shock protein B6 | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heat shock protein beta-6, ... | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
8JWY
 
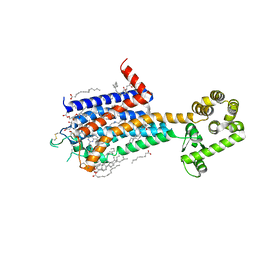 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
