8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QMO
 
 | | Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Kwong, H.S, Grandvuillemin, L, Sirounian, S, Ancelin, A, Lai-Kee-Him, J, Carivenc, C, Lancey, C, Ragan, T.J, Hesketh, E.L, Bourguet, W, Gruszczyk, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
J.Mol.Biol., 436, 2024
|
|
7Z6L
 
 | | Crystal structure of PROTAC 5 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[3-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]propoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Roy, M.J, Bader, G, Farnaby, W, Ciulli, A. | | Deposit date: | 2022-03-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
9DIP
 
 | | Crystal structure of H5 hemagglutinin from the influenza virus A/Texas/37/2024 (H5N1) with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-09-05 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A single mutation in bovine influenza H5N1 hemagglutinin switches specificity to human receptors.
Science, 386, 2024
|
|
6OC3
 
 | |
6GH7
 
 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[(3~{R})-pyrrolidin-3-yl]pentanamide, Streptavidin | | Authors: | Nodling, A.R, Tsai, Y.H, Luk, L.Y.P, Rizkallah, P, Jin, Y. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Reactivity and Selectivity of Iminium Organocatalysis Improved by a Protein Host.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
9EZG
 
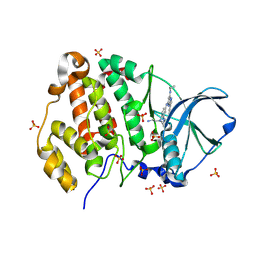 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((4-((2-aminoethyl)(ethyl)amino)-3-(4H-1,2,4-triazol-4-yl)phenyl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 5-[[4-[2-azanylethyl(ethyl)amino]-3-(1,2,4-triazol-4-yl)phenyl]amino]-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
8P07
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((3-(4H-1,2,4-triazol-4-yl)phenyl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-[[3-(1,2,4-triazol-4-yl)phenyl]amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Bown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
8P06
 
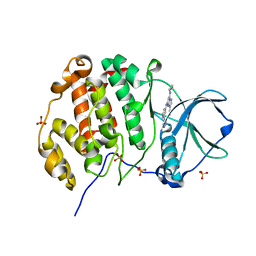 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((2-(4H-1,2,4-triazol-4-yl)pyridin-4-yl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 7-(cyclopropylamino)-5-[[2-(1,2,4-triazol-4-yl)pyridin-4-yl]amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
8CLH
 
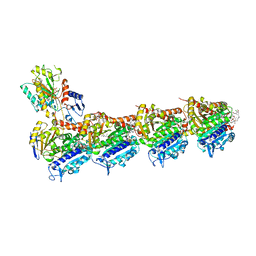 | | Drug cocktail (Colchicine, Epothilone A, Peloruside, Ansamitocin P3, Vinblastine) bound to tubulin (T2R-TTL) complex | | Descriptor: | (1S,2S,3S,5S,6S,16Z,18Z,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl 2-methylpropanoate, (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, CALCIUM ION, ... | | Authors: | Wranik, M, Bertrand, Q, Kepa, M.W, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
5KDU
 
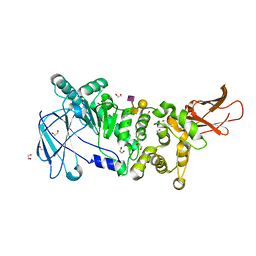 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDN
 
 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, ZINC ION | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8PJE
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAT | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Hesketh, S.J, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
8PJF
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
8P9W
 
 | | vitamin D receptor complex with Xe4MeCF3 analog | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-1,7~{a}-dimethyl-1-[6,6,6-tris(fluoranyl)-5-oxidanyl-5-(trifluoromethyl)hexa-1,3-diynyl]-2,3,3~{a},5,6,7-hexahydroinden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Belorusova, A.Y, Rochel, N. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A vitamin D-based strategy overcomes chemoresistance in prostate cancer.
Br.J.Pharmacol., 181, 2024
|
|
5KDS
 
 | | ZmpB metallopeptidase in complex with an O-glycopeptide (a2,6-sialylated core-3 pentapeptide). | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KF2
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 8 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Holden, H.M, Thoden, J.B, Dopkins, B.J, Tipton, P.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
3DOG
 
 | | Structure of Thr 160 phosphorylated CDK2/cyclin A in complex with the inhibitor N-&-N1 | | Descriptor: | (2R)-2-{[4-(benzylamino)-8-(1-methylethyl)pyrazolo[1,5-a][1,3,5]triazin-2-yl]amino}butan-1-ol, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Endicott, J. | | Deposit date: | 2008-07-04 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-&-N, a new class of cell death-inducing kinase inhibitors derived from the purine roscovitine.
Mol.Cancer Ther., 7, 2008
|
|
4XZ3
 
 | |
7QNE
 
 | | Cryo-EM structure of human full-length synaptic alpha1beta3gamma2 GABA(A)R in complex with Ro15-4513 and megabody Mb38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
9DIO
 
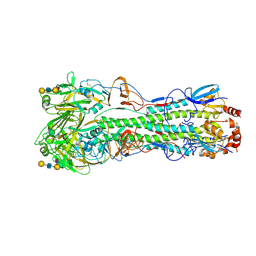 | | Crystal structure of H5 hemagglutinin Q226L mutant from the influenza virus A/Texas/37/2024 (H5N1) with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-09-05 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A single mutation in bovine influenza H5N1 hemagglutinin switches specificity to human receptors.
Science, 386, 2024
|
|
8OLT
 
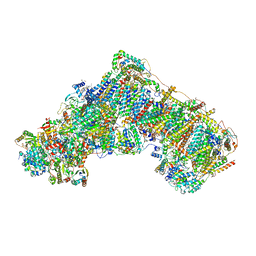 | | Mitochondrial complex I from Mus musculus in the active state bound with piericidin A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
9EH5
 
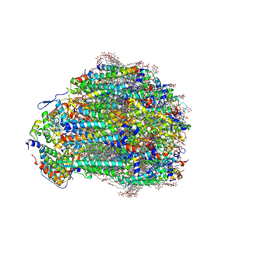 | | Structure of a mutated photosystem II complex reveals changes to the hydrogen-bonding network that affect proton egress during O-O bond formation | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Flesher, D.A, Shin, J, Debus, R.J, Brudvig, G.W. | | Deposit date: | 2024-11-22 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | Structure of a mutated photosystem II complex reveals changes to the hydrogen-bonding network that affect proton egress during O-O bond formation.
J.Biol.Chem., 301, 2025
|
|
8FC5
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
