7U1D
 
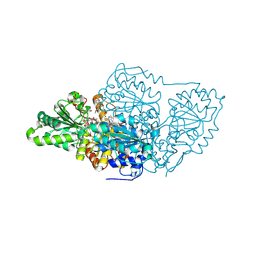 | |
7TZZ
 
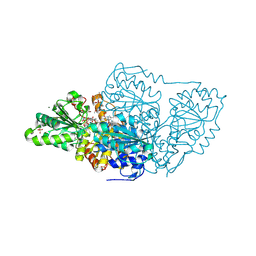 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase P197T mutant in complex with bispyribac-sodium | | Descriptor: | 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
7TOS
 
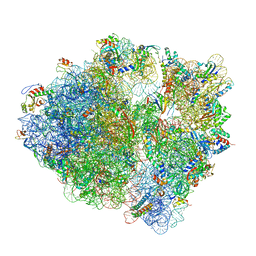 | | E. coli 70S ribosomes bound with the ALS/FTD-associated dipeptide repeat protein PR20 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
7U1N
 
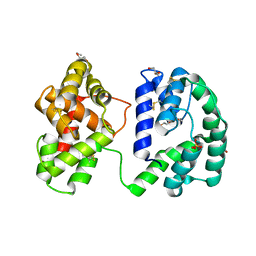 | | Crystal structure of the Anopheles darlingi AD-118 long form D7 salivary protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-ETHOXYETHANOL, ... | | Authors: | Alvarenga, P.H, Gittis, A.G, Garboczi, D.N, Andersen, J.F. | | Deposit date: | 2022-02-21 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional aspects of evolution in a cluster of salivary protein genes from mosquitoes.
Insect Biochem.Mol.Biol., 146, 2022
|
|
2C64
 
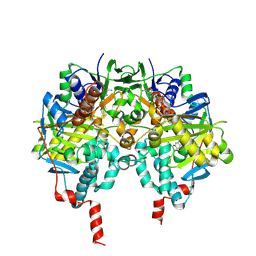 | | MAO inhibition by rasagiline analogues | | Descriptor: | (1R)-6-HYDROXY-N-METHYL-N-[(1Z)-PROP-2-EN-1-YLIDENE]INDAN-1-AMINIUM, AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of Rasagiline-Related Inhibitors to Human Monoamine Oxidases: A Kinetic and Crystallographic Analysis.
J.Med.Chem., 48, 2005
|
|
2BRD
 
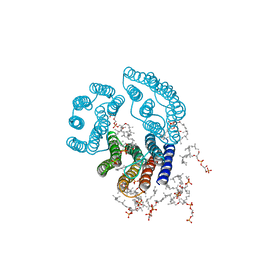 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN PURPLE MEMBRANE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Henderson, R, Grigorieff, N. | | Deposit date: | 1995-12-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Electron-crystallographic refinement of the structure of bacteriorhodopsin.
J.Mol.Biol., 259, 1996
|
|
7TSD
 
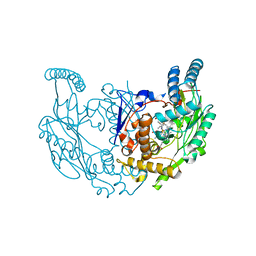 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with 6-(3-(3,3-difluoroazetidin-1-yl)prop-1-yn-1-yl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[3-(3,3-difluoroazetidin-1-yl)prop-1-yn-1-yl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | 2-Aminopyridines with a shortened amino sidechain as potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem., 69, 2022
|
|
2BZG
 
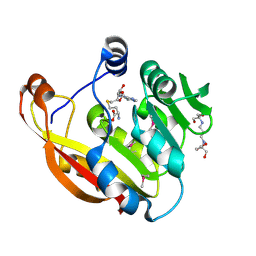 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
5FWG
 
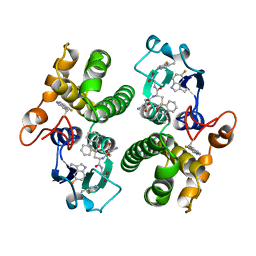 | | TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE MU CLASS | | Authors: | Parsons, J.F, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-11-08 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes harboring unnatural amino acids: mechanistic and structural analysis of the enhanced catalytic activity of a glutathione transferase containing 5-fluorotryptophan.
Biochemistry, 37, 1998
|
|
2I42
 
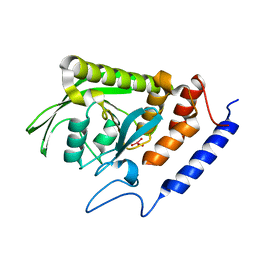 | |
7TUR
 
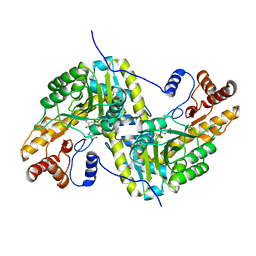 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
2I5Q
 
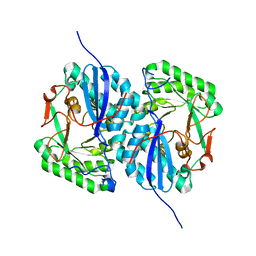 | | Crystal structure of Apo L-rhamnonate dehydratase from Escherichia Coli | | Descriptor: | L-rhamnonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
2C0U
 
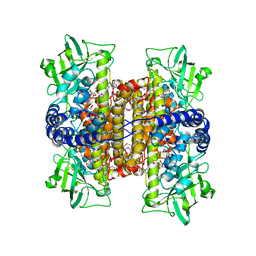 | | Crystal Structure of a Covalent Complex of Nitroalkane Oxidase Trapped During Substrate Turnover | | Descriptor: | (2S)-2-NITROBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, NITROALKANE OXIDASE | | Authors: | Nagpal, A, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2005-09-07 | | Release date: | 2006-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Nitroalkane Oxidase: Insights Into the Reaction Mechanism from a Covalent Complex of the Flavoenzyme Trapped During Turnover.
Biochemistry, 45, 2006
|
|
2I68
 
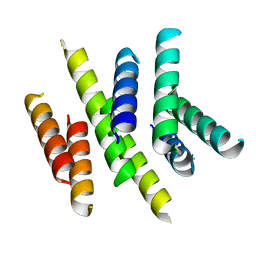 | | Cryo-EM based theoretical model structure of transmembrane domain of the multidrug-resistance antiporter from E. coli EmrE | | Descriptor: | Protein emrE | | Authors: | Fleishman, S.J, Harrington, S.E, Enosh, A, Halperin, D, Tate, C.G, Ben-Tal, N. | | Deposit date: | 2006-08-28 | | Release date: | 2006-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (7.5 Å) | | Cite: | Quasi-symmetry in the Cryo-EM Structure of EmrE Provides the Key to Modeling its Transmembrane Domain
J.Mol.Biol., 364, 2006
|
|
7TS3
 
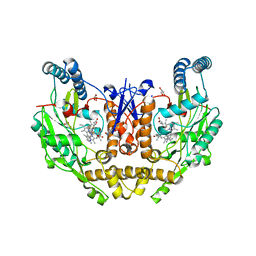 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-(3,3-difluoroazetidin-1-yl)prop-1-yn-1-yl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[3-(3,3-difluoroazetidin-1-yl)prop-1-yn-1-yl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | 2-Aminopyridines with a shortened amino sidechain as potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem., 69, 2022
|
|
2CAB
 
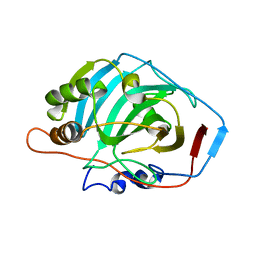 | | STRUCTURE, REFINEMENT AND FUNCTION OF CARBONIC ANHYDRASE ISOZYMES. REFINEMENT OF HUMAN CARBONIC ANHYDRASE I | | Descriptor: | CARBONIC ANHYDRASE FORM B, ZINC ION | | Authors: | Kannan, K.K, Ramanadham, M, Jones, T.A. | | Deposit date: | 1983-10-05 | | Release date: | 1984-02-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, refinement, and function of carbonic anhydrase isozymes: refinement of human carbonic anhydrase I
Ann.N.Y.Acad.Sci., 429, 1984
|
|
2CB8
 
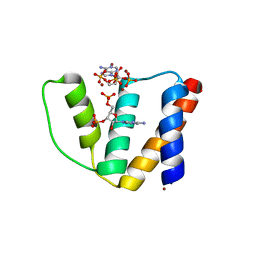 | | High resolution crystal structure of liganded human L-ACBP | | Descriptor: | 2-METHOXYETHANOL, ACYL-COA-BINDING PROTEIN, SULFATE ION, ... | | Authors: | Taskinen, J.P, van Aalten, D.M, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2006-01-03 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Crystal Structures of Unliganded and Liganded Human Liver Acbp Reveal a New Mode of Binding for the Acyl-Coa Ligand.
Proteins: Struct., Funct., Bioinf., 66, 2007
|
|
2CBC
 
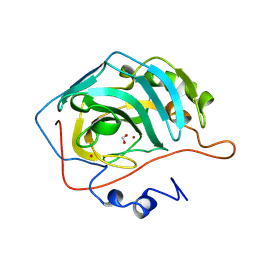 | | STRUCTURE OF NATIVE AND APO CARBONIC ANHYDRASE II AND SOME OF ITS ANION-LIGAND COMPLEXES | | Descriptor: | CARBONIC ANHYDRASE II, FORMIC ACID, MERCURY (II) ION, ... | | Authors: | Hakansson, K, Carlsson, M, Svensson, L.A, Liljas, A. | | Deposit date: | 1992-06-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes.
J.Mol.Biol., 227, 1992
|
|
2CBI
 
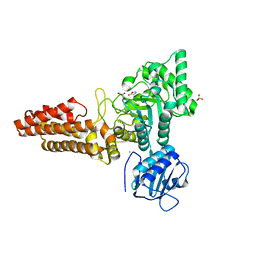 | | Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase | | Descriptor: | CHLORIDE ION, GAMMA-BUTYROLACTONE, GLYCEROL, ... | | Authors: | Rao, F.V, Dorfmueller, H.C, Villa, F, Allwood, M, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2006-01-04 | | Release date: | 2006-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis.
EMBO J., 25, 2006
|
|
2HZK
 
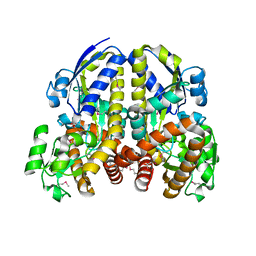 | | Crystal structures of a sodium-alpha-keto acid binding subunit from a TRAP transporter in its open form | | Descriptor: | GLYCEROL, TRAP-T family sorbitol/mannitol transporter, periplasmic binding protein, ... | | Authors: | Gonin, S, Arnoux, P, Pierru, B, Alonso, B, Sabaty, M, Pignol, D. | | Deposit date: | 2006-08-09 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding.
Bmc Struct.Biol., 7, 2007
|
|
2CBB
 
 | | STRUCTURE OF NATIVE AND APO CARBONIC ANHYDRASE II AND SOME OF ITS ANION-LIGAND COMPLEXES | | Descriptor: | CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Hakansson, K, Carlsson, M, Svensson, L.A, Liljas, A. | | Deposit date: | 1992-06-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes.
J.Mol.Biol., 227, 1992
|
|
7TM3
 
 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TDD
 
 | | AtTPC1 D454N-EDTA state II | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TDE
 
 | | AtTPC1 DDE mutant with 1 mM Ca2+ | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TUT
 
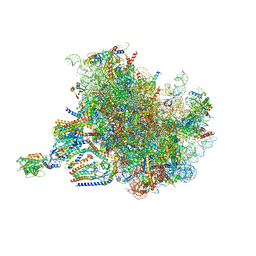 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
