3W5P
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
3W5Q
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (5beta,9beta)-3-oxocholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
5M2H
 
 | | Crystal structure of vancomycin-Zn(II)-citrate complex | | Descriptor: | CITRIC ACID, HEXAETHYLENE GLYCOL, Vancomycin, ... | | Authors: | Zarkan, A, Macklyne, H.-R, Chirgadze, D.Y, Bond, A.D, Hesketh, A.R, Hong, H.-J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Sci Rep, 7, 2017
|
|
2P0D
 
 | | ArhGAP9 PH domain in complex with Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-04-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4LE7
 
 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
1PV1
 
 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | Descriptor: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | Authors: | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-26 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
5U9T
 
 | | The Tris-thiolate Zn(II)S3Cl Binding Site Engineered by D-Cysteine Ligands in de Novo Three-stranded Coiled Coil Environment | | Descriptor: | ACETATE ION, CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Ruckthong, L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | d-Cysteine Ligands Control Metal Geometries within De Novo Designed Three-Stranded Coiled Coils.
Chemistry, 23, 2017
|
|
8P6Q
 
 | | Racemic structure of TNFR1 cysteine-rich domain | | Descriptor: | D-TNFR-1 CRD2, SULFATE ION, Tumor necrosis factor-binding protein 1 | | Authors: | Lander, A.J, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2023-05-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deciphering the Synthetic and Refolding Strategy of a Cysteine-Rich Domain in the Tumor Necrosis Factor Receptor (TNF-R) for Racemic Crystallography Analysis and d-Peptide Ligand Discovery.
Acs Bio Med Chem Au, 4, 2024
|
|
6EXP
 
 | | Crystal structure of the SIRV3 AcrID1 (gp02) anti-CRISPR protein | | Descriptor: | SIRV3 AcrID1 (gp02) anti-CRISPR protein | | Authors: | He, F, Bhoobalan-Chitty, Y, Van, L.B, Kjeldsen, A.L, Dedola, M, Makarova, K.S, Koonin, E.V, Brodersen, D.E, Peng, X. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
Nat Microbiol, 3, 2018
|
|
6F9J
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-mannopyranose-(1-4)-1-DEOXYNOJIRIMYCIN | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
4H2Z
 
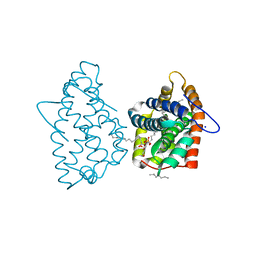 | | Crystal structure of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE, ... | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-09-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4PKR
 
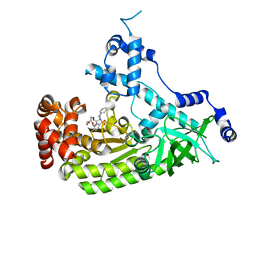 | | Anthrax toxin lethal factor with bound small molecule inhibitor 10 | | Descriptor: | CHLORIDE ION, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1S2J
 
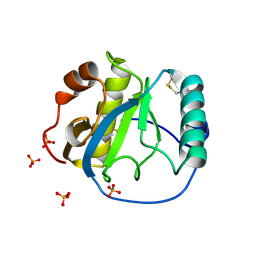 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
4KSH
 
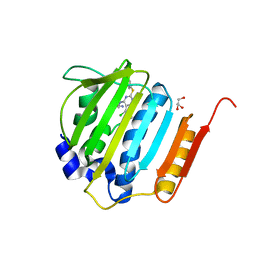 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (7-({4-[(3R)-3-AMINOPYRROLIDIN-1-YL]-5-CHLORO-6-ETHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-2-YL}SULFANYL)-1,5-NAPHTHYRIDIN-1(4H)-OL) | | Descriptor: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1,5-naphthyridin-1(4H)-ol, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | Deposit date: | 2013-05-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with broad-spectrum antibacterial activity
To be Published
|
|
1D31
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF BULGE-CONTAINING DNA FRAGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Joshua-Tor, L, Frolow, F, Appella, E, Hope, H, Rabinovich, D, Sussman, J.L. | | Deposit date: | 1991-04-25 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of bulge-containing DNA fragments.
J.Mol.Biol., 225, 1992
|
|
9FFM
 
 | |
9FF2
 
 | |
9FEZ
 
 | |
4XAK
 
 | | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain of neutralizing antibody m336, ... | | Authors: | Zhou, T, Dimtrov, D.S, Ying, T. | | Deposit date: | 2014-12-15 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Nat Commun, 6, 2015
|
|
9FFN
 
 | |
5OX4
 
 | | Glycogen Phosphorylase in complex with CK900 | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[5-(4-aminophenyl)-4~{H}-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
3NU0
 
 | | Design, Synthesis, Biological Evaluation and X-ray Crystal Structure of Novel Classical 6,5,6-TricyclicBenzo[4,5]thieno[2,3-d]pyrimidines as Dual Thymidylate Synthase and Dihydrofolate Reductase Inhibitors | | Descriptor: | (2S)-2-(5-{[(2-amino-4-oxo-3,4-dihydro[1]benzothieno[2,3-d]pyrimidin-5-yl)methyl]amino}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)pentanedioic acid, Dihydrofolate reducatase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
8FFF
 
 | |
6J4H
 
 | | Crystal Structure of maltotriose-complex of PulA-G680L mutant from Klebsiella pneumoniae | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
