6HS7
 
 | | Type VI membrane complex | | Descriptor: | ImcF-like family protein, Type VI secretion system protein VasD | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | In situand high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex.
Embo J., 38, 2019
|
|
3T2J
 
 | | Tetragonal thermolysin in the presence of betaine | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cahn, J, Hti Lar Seng, N.S, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SZE
 
 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | Descriptor: | Serine protease espP | | Authors: | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
6HD0
 
 | |
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3T3R
 
 | | Human Cytochrome P450 2A6 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
6HVU
 
 | |
6JPQ
 
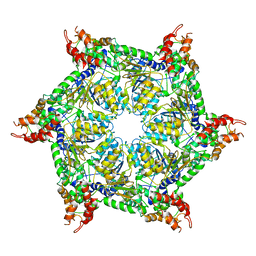 | | CryoEM structure of Abo1 hexamer - ADP complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6JDX
 
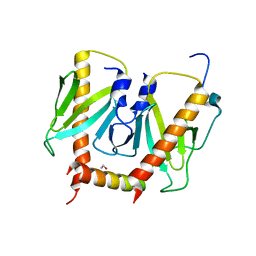 | | Crystal structure of AcrIIC2 dimer in complex with partial Nme1Cas9 preprocessed with protease alpha-Chymotrypsin | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Cheng, Z, Huang, X, Sun, W, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
6MKS
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | Chimera protein of NLR family CARD domain-containing protein 4 and EGFP | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NLRC4CARDfilament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly.
J. Biol. Chem., 293, 2018
|
|
6JMH
 
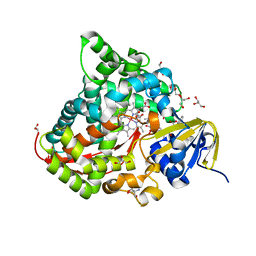 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JMT
 
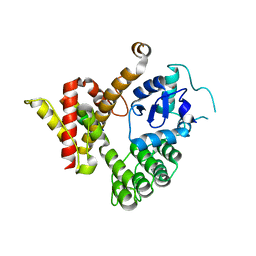 | | Crystal structure of GIT/PIX complex | | Descriptor: | ARF GTPase-activating protein GIT2, ZINC ION, beta PIX | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
6MJL
 
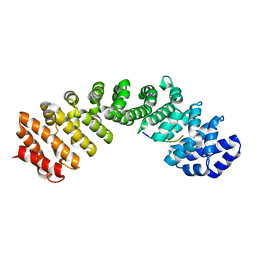 | | Crystal structure of ChREBP NLS peptide bound to importin alpha. | | Descriptor: | ChREBP Peptide ASN-TYR-TRP-LYS-ARG-ARG-ILE-GLU-VAL, Importin subunit alpha-1 | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of importin alpha and the nuclear localization peptide of ChREBP, and small compound inhibitors of ChREBP-importin alpha interactions.
Biochem.J., 477, 2020
|
|
6MKR
 
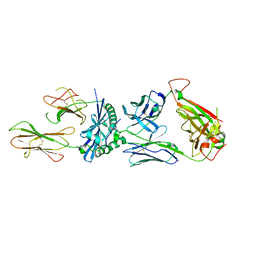 | | 5287 TCR bound to IAb Padi4 | | Descriptor: | 5287 TCR alpha chain, 5287 TCR beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Blevins, S.J, Huseby, E.S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.355 Å) | | Cite: | A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+Tregcell development.
Nat.Immunol., 20, 2019
|
|
6JP5
 
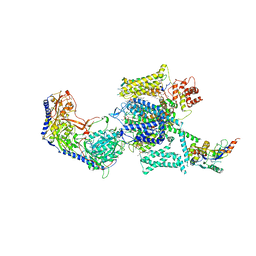 | | Rabbit Cav1.1-Nifedipine Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6MK9
 
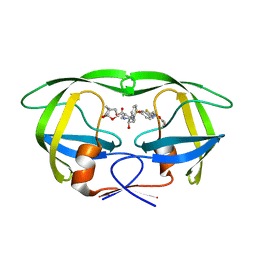 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121
To Be Published
|
|
3S7S
 
 | |
3S82
 
 | |
6J7I
 
 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (15aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
3SHF
 
 | |
6GSA
 
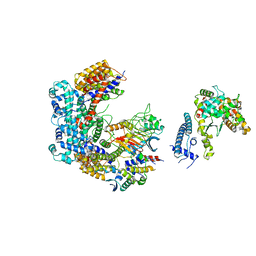 | | Core Centromere Binding Factor 3 (CBF3) with monomeric Ndc10 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Zhang, W.J, Lukoynova, N, Vaughan, C.K. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
6GTZ
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound dimannoside Man(alpha1-3)Man in space group P21 | | Descriptor: | FimG protein, FimH protein, alpha-D-mannopyranose-(1-3)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GYP
 
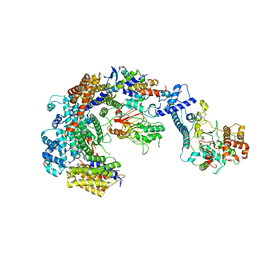 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3SYX
 
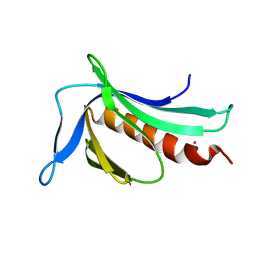 | | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein. Northeast Structural Genomics Consortium Target HR5538B. | | Descriptor: | Sprouty-related, EVH1 domain-containing protein 1, YTTRIUM (III) ION | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein.
To be Published
|
|
