4ACM
 
 | | CDK2 IN COMPLEX WITH 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}-PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE | | Descriptor: | 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2, GLYCEROL | | Authors: | Berg, S, Bhat, R, Anderson, M, Bergh, M, Brassington, C, Hellberg, S, Jerning, E, Hogdin, K, Lo-Alfredsson, Y, Neelissen, J, Nilsson, Y, Ormo, M, Soderman, P, Stanway, J, Tucker, J, von Berg, S, Weigelt, T, Xue, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
6GHK
 
 | | Human PARP1 (ARTD1) - Catalytic domain in complex with inhibitor ME0527 | | Descriptor: | Poly [ADP-ribose] polymerase 1, SULFATE ION, ~{N}-[(1~{R})-1-(4-imidazol-1-ylphenyl)ethyl]-3-(4-oxidanylidene-1~{H}-quinazolin-2-yl)propanamide | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Moche, M, Brock, J, Ekblad, T, Spjut, S, Elofsson, M, Schuler, H. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human PARP1 (ARTD1) - Catalytic domain in complex with inhibitor ME0527
To Be Published
|
|
6GTT
 
 | | Human STK10 bound to BIRB-796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Serine/threonine-protein kinase 10 | | Authors: | Berger, B.T, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | STK10 bound to BIRB-796
To Be Published
|
|
7DRZ
 
 | |
7DS0
 
 | |
7DS1
 
 | | Crystal structure of Aspergillus oryzae Rib2 deaminase in complex with DARIPP (C-terminal deletion mutant at pH 6.5) | | Descriptor: | CMP/dCMP-type deaminase domain-containing protein, ZINC ION, [(2~{R},3~{S},4~{S})-5-[[2,5-bis(azanyl)-6-oxidanylidene-1~{H}-pyrimidin-4-yl]amino]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Chen, S.C, Liaw, S.H, Hsu, C.H. | | Deposit date: | 2020-12-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of Aspergillus oryzae Rib2 deaminase: the functional mechanism involved in riboflavin biosynthesis.
Iucrj, 8, 2021
|
|
7DRY
 
 | | Crystal structure of Aspergillus oryzae Rib2 deaminase | | Descriptor: | CMP/dCMP-type deaminase domain-containing protein, SULFATE ION, ZINC ION | | Authors: | Chen, S.C, Liaw, S.H, Hsu, C.H. | | Deposit date: | 2020-12-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of Aspergillus oryzae Rib2 deaminase: the functional mechanism involved in riboflavin biosynthesis.
Iucrj, 8, 2021
|
|
3SMJ
 
 | | Human poly(ADP-ribose) polymerase 14 (Parp14/Artd8) - catalytic domain in complex with a pyrimidine-like inhibitor | | Descriptor: | 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, GLYCEROL, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
3SE2
 
 | | Human poly(ADP-ribose) polymerase 14 (PARP14/ARTD8) - catalytic domain in complex with 6(5H)-phenanthridinone | | Descriptor: | 3-aminobenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Siponen, M.I, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
3UF0
 
 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
4ARB
 
 | | Mus musculus Acetylcholinesterase in complex with (S)-C5685 at 2.25 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ACETYLCHOLINESTERASE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4ARA
 
 | | Mus musculus Acetylcholinesterase in complex with (R)-C5685 at 2.5 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3SMI
 
 | | Human poly(ADP-ribose) polymerase 14 (Parp14/Artd8) - catalytic domain in complex with a quinazoline inhibitor | | Descriptor: | 2-{[(3-amino-1H-1,2,4-triazol-5-yl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
4ATI
 
 | | MITF:M-box complex | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*CP*AP*TP*GP*TP*GP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*CP*AP*CP*AP*TP*GP*AP*CP*CP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | Authors: | Pogenberg, V, Deineko, V, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
4ATH
 
 | | MITF apo structure | | Descriptor: | MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
3QPD
 
 | | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon | | Descriptor: | Cutinase 1 | | Authors: | Lu, A, Gosser, Y, Montclare, J.K, Liu, Z, Kong, X. | | Deposit date: | 2011-02-11 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon
To be Published
|
|
3ZME
 
 | |
4NDI
 
 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDF
 
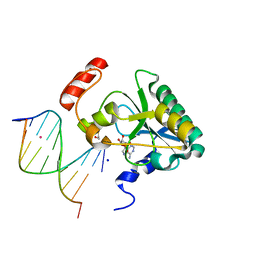 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
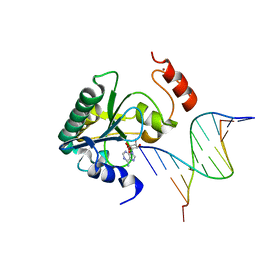 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NPR
 
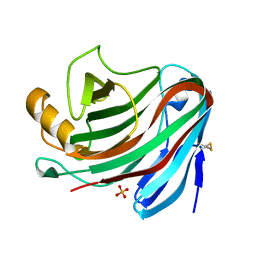 | | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase GH12 | | Authors: | Cordeiro, R.L, Santos, C.R, Furtado, G.P, Damasio, A.R.L, Polizeli, M.L.T.M, Ward, R.J, Murakami, M.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus
To be Published
|
|
5TNC
 
 | |
4G08
 
 | |
4J1A
 
 | |
