7QP6
 
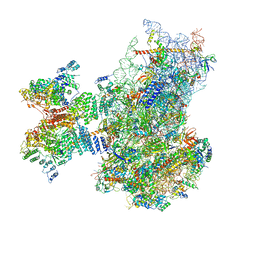 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
6N52
 
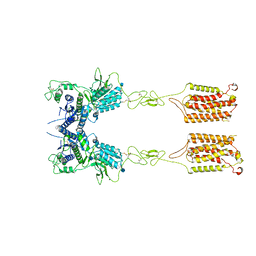 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
8STD
 
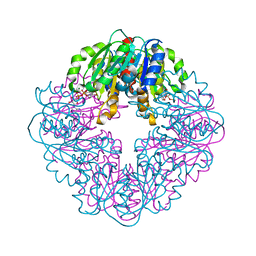 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD and soaked with CS2 | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
8SOQ
 
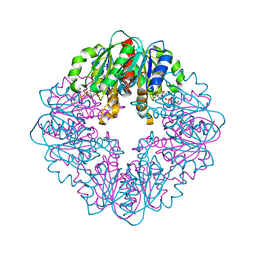 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
7UB2
 
 | |
6WVJ
 
 | | Cryo-EM structure of Bacillus subtilis RNA Polymerase elongation complex | | Descriptor: | DNA (5'-D(*TP*GP*TP*CP*GP*GP*GP*CP*GP*TP*CP*CP*GP*CP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*AP*CP*GP*CP*CP*CP*GP*AP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Newing, T, Tolun, G, Oakley, A.J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.
Nat Commun, 11, 2020
|
|
6VZ4
 
 | |
6MUO
 
 | |
7YU5
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU3
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU8
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU6
 
 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state2 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
6WVK
 
 | | Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex with HelD | | Descriptor: | DNA helicase IV, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Newing, T, Tolun, G, Oakley, A.J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.
Nat Commun, 11, 2020
|
|
6X2L
 
 | | hEAAT3-IFS-Na | | Descriptor: | Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
6X2Z
 
 | | hEAAT3-OFS-Asp | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
6X3E
 
 | | hEAAT3-Asymmetric-1o2i | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
6X3F
 
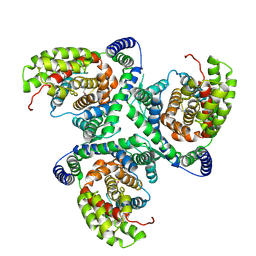 | | hEAAT3-IFS-Apo | | Descriptor: | CHOLINE ION, Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
6S5P
 
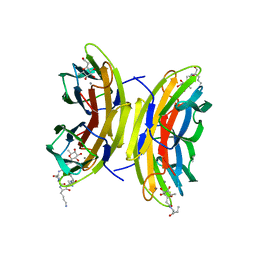 | | Cfucosylated peptide SBL2 bound to Fucose binding Lectin LecB (PA-IIL) from Pseudomonas aeruginosa at 1.46 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
7YU7
 
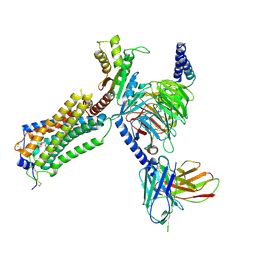 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state3 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
8SR7
 
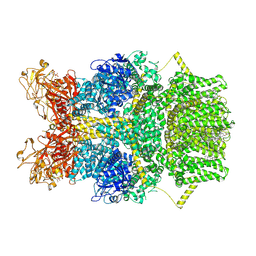 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
6NMN
 
 | |
8R1O
 
 | | Structure of C. thermophilum RNA exosome core | | Descriptor: | Exoribonuclease phosphorolytic domain-containing protein, Exoribonuclease-like protein, Exosome complex component MTR3, ... | | Authors: | Lazzaretti, D, Liebau, J, Pilsl, M, Sprangers, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Beyond static structures: quantitative dynamics in the eukaryotic RNA exosome complex
Biorxiv, 2024
|
|
8ANW
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8). | | Descriptor: | Capsid protein, VP0, VP1, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and Characterisation of Stabilised PV-3 Virus-like Particles Using Pichia pastoris .
Viruses, 14, 2022
|
|
8T2J
 
 | | Structure of the catalytic domain of PPM1D/Wip1 serine/threonine phosphatase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumar, J.P, Kosek, D, Dyda, F. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanistic studies of the PPM1D serine/threonine phosphatase catalytic domain.
J.Biol.Chem., 300, 2024
|
|
8FHM
 
 | | RNase A-Uridine 5'-Hexaphosphate (RNaseA.p6U) | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]uridine, Ribonuclease pancreatic | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-08-07 | | Method: | SOLUTION SCATTERING (1.79 Å), X-RAY DIFFRACTION | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A.
Acs Cent.Sci., 10, 2024
|
|
