1DVN
 
 | |
3EOX
 
 | |
3EVJ
 
 | | Intermediate structure of antithrombin bound to the natural pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Huntington, J.A, Belzar, K.J. | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The critical role of hinge-region expulsion in the induced-fit heparin binding mechanism of antithrombin.
J. Mol. Biol., 386, 2009
|
|
1F0C
 
 | | STRUCTURE OF THE VIRAL SERPIN CRMA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ICE INHIBITOR | | Authors: | Renatus, M, Zhou, Q, Stennicke, H.R, Snipas, S.J, Turk, D, Bankston, L.A, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the apoptotic suppressor CrmA in its cleaved form.
Structure Fold.Des., 8, 2000
|
|
1E04
 
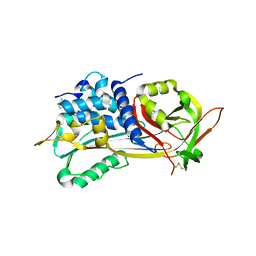 | | PLASMA BETA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, GLYCEROL, ... | | Authors: | Mccoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1DVM
 
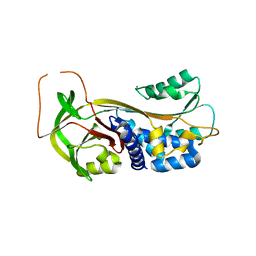 | | ACTIVE FORM OF HUMAN PAI-1 | | Descriptor: | CHLORIDE ION, PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Stout, T.J, Graham, H, Buckley, D.I, Matthews, D.J. | | Deposit date: | 2000-01-21 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of active and latent PAI-1: a possible stabilizing role for chloride ions.
Biochemistry, 39, 2000
|
|
3F02
 
 | |
1E03
 
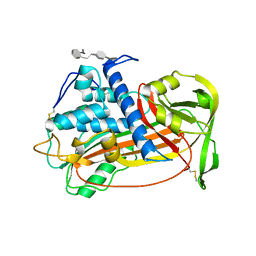 | | PLASMA ALPHA ANTITHROMBIN-III AND PENTASACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ... | | Authors: | McCoy, A.J, Jin, L, Abrahams, J.-P, Skinner, R, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1DZH
 
 | | P14-FLUORESCEIN-N135Q-S380C-ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Mccoy, A.J, Huntington, J.A, Carrell, R.W. | | Deposit date: | 2000-02-28 | | Release date: | 2000-05-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Conformational Activation of Antithrombin. A 2. 85-A Structure of a Fluorescein Derivative Reveals an Electrostatic Link between the Hinge and Heparin Binding Regions.
J.Biol.Chem., 275, 2000
|
|
1E05
 
 | | PLASMA ALPHA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | McCoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1DZG
 
 | | N135Q-S380C-ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | McCoy, A.J, Huntington, J.A, Carrell, R.W. | | Deposit date: | 2000-02-28 | | Release date: | 2000-05-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Conformational Activation of Antithrombin. A 2. 85-A Structure of a Fluorescein Derivative Reveals an Electrostatic Link between the Hinge and Heparin Binding Regions.
J.Biol.Chem., 275, 2000
|
|
3DRM
 
 | | 2.2 Angstrom Crystal Structure of Thr114Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
3CWL
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
3DY0
 
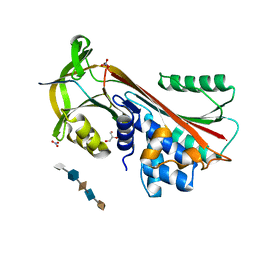 | | Crystal Structure of Cleaved PCI Bound to Heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-terminus Plasma serine protease inhibitor, GLYCEROL, ... | | Authors: | Li, W, Huntington, J.A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The heparin binding site of protein C inhibitor is protease-dependent.
J.Biol.Chem., 283, 2008
|
|
3DRU
 
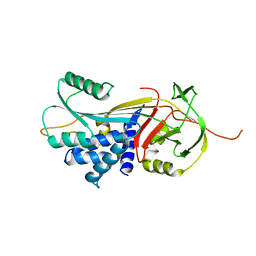 | | Crystal Structure of Gly117Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
3DLW
 
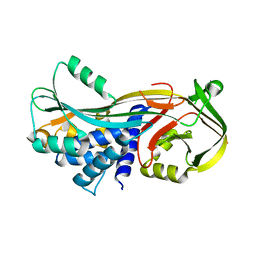 | | Antichymotrypsin | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Feil, S.C. | | Deposit date: | 2008-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and characterization of a misfolded monomeric serpin formed at physiological temperature
J.Mol.Biol., 403, 2010
|
|
3F5N
 
 | |
1DB2
 
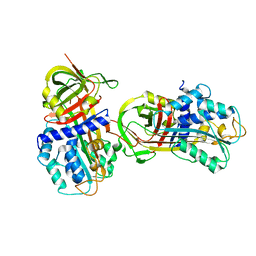 | | CRYSTAL STRUCTURE OF NATIVE PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Nar, H, Bauer, M, Stassen, J.M, Lang, D, Gils, A, Declerck, P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation.
J.Mol.Biol., 297, 2000
|
|
3FGQ
 
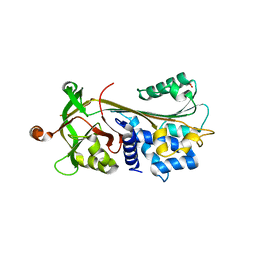 | | Crystal structure of native human neuroserpin | | Descriptor: | GLYCEROL, Neuroserpin | | Authors: | Takehara, S, Yang, X, Mikami, B, Onda, M. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability
J.Mol.Biol., 388, 2009
|
|
3CWM
 
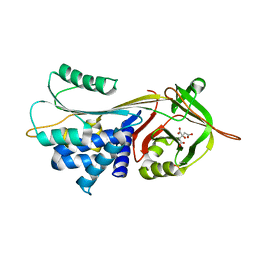 | | Crystal structure of alpha-1-antitrypsin complexed with citrate | | Descriptor: | Alpha-1-antitrypsin, CITRIC ACID | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
1HP7
 
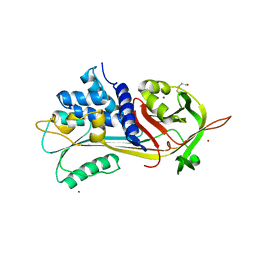 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | Descriptor: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | Authors: | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
1HLE
 
 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
1IMV
 
 | | 2.85 A crystal structure of PEDF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PIGMENT EPITHELIUM-DERIVED FACTOR | | Authors: | Simonovic, M, Gettins, P.G.W, Volz, K. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1AS4
 
 | | CLEAVED ANTICHYMOTRYPSIN A349R | | Descriptor: | ACETATE ION, ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1IZ2
 
 | | Interactions causing the kinetic trap in serpin protein folding | | Descriptor: | alpha-D-glucopyranose-(1-2)-(5R)-5-[(2R)-2-hydroxynonyl]-beta-D-xylulofuranose, alpha1-antitrypsin | | Authors: | Im, H, Woo, M.-S, Hwang, K.Y, Yu, M.-H. | | Deposit date: | 2002-09-19 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions causing the kinetic trap in serpin protein folding
J.BIOL.CHEM., 277, 2002
|
|
